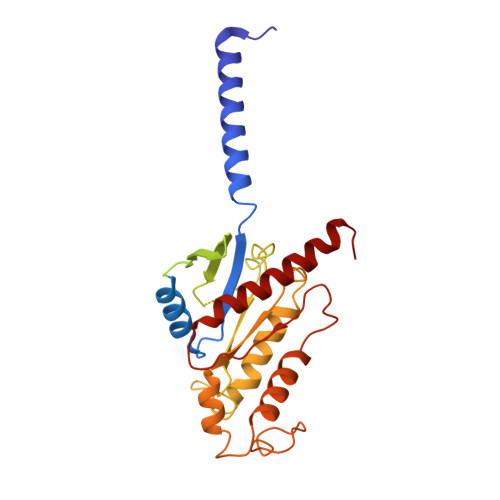
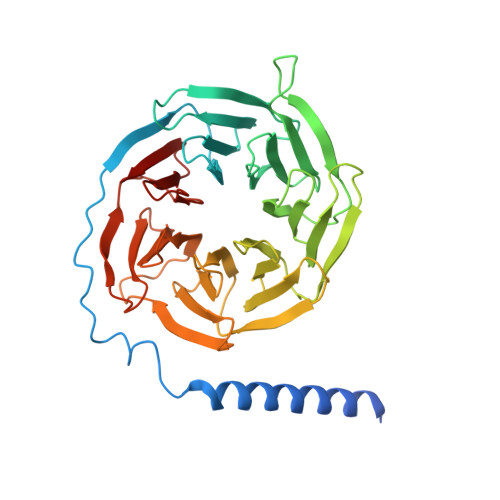
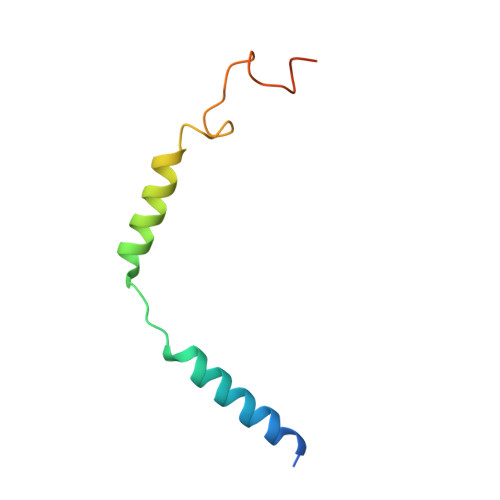
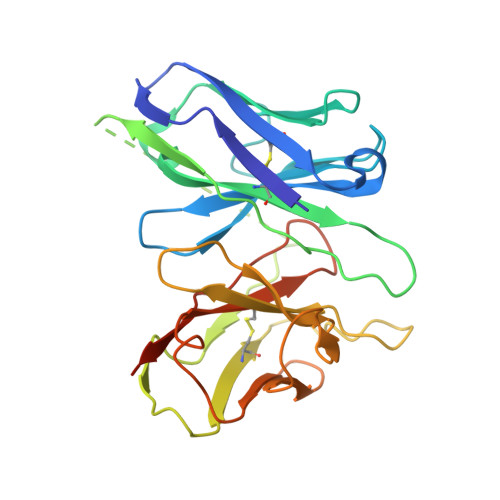
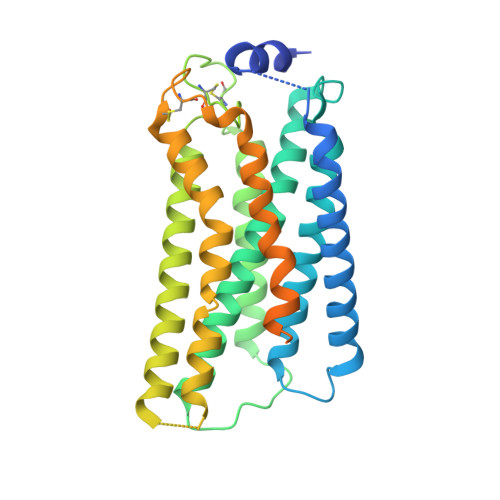
Structure of S1PR2-heterotrimeric G 13 signaling complex.
Chen, H., Chen, K., Huang, W., Staudt, L.M., Cyster, J.G., Li, X.(2022) Sci Adv 8: eabn0067-eabn0067
- PubMed: 35353559
- DOI: https://doi.org/10.1126/sciadv.abn0067
- Primary Citation of Related Structures:
7T6B - PubMed Abstract:
Sphingosine-1-phosphate (S1P) regulates immune cell trafficking, angiogenesis, and vascular function via its five receptors. Inherited mutations in S1P receptor 2 (S1PR2) occur in individuals with hearing loss, and acquired mutations in S1PR2 and G α13 occur in a malignant lymphoma. Here, we present the cryo-electron microscopy structure of S1P-bound S1PR2 coupled to the heterotrimeric G 13 . Interaction between S1PR2 intracellular loop 2 (ICL2) and transmembrane helix 4 confines ICL2 to engage the α5 helix of G α13 . Transforming growth factor-α shedding assays and cell migration assays support the key roles of the residues in S1PR2-G α13 complex assembly. The structure illuminates the mechanism of receptor disruption by disease-associated mutations. Unexpectedly, we showed that FTY720-P, an agonist of the other four S1PRs, can trigger G 13 activation via S1PR2. S1PR2 F274I variant can increase the activity of G 13 considerably with FTY720-P and S1P, thus revealing a basis for S1PR drug selectivity.
- Department of Molecular Genetics, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: