Sequence of conformation changes of DnaB helicase during DNA unwinding and priming in Escherichia coli
Oakley, A.J., Xu, Z.Q.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replicative DNA helicase | 471 | Escherichia coli K-12 | Mutation(s): 1 Gene Names: dnaB, groP, grpA, b4052, JW4012 EC: 3.6.4.12 (PDB Primary Data), 5.6.2.3 (UniProt) | 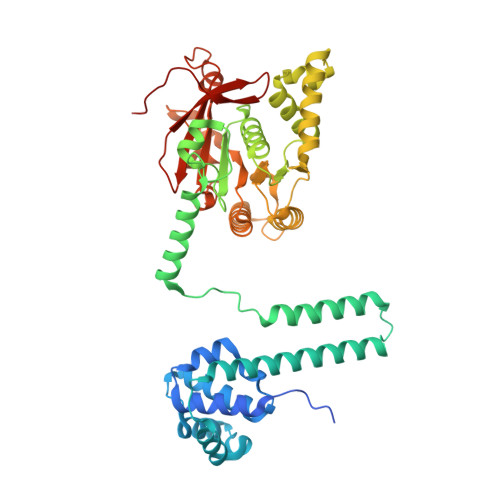 | |
UniProt | |||||
Find proteins for P0ACB0 (Escherichia coli (strain K12)) Explore P0ACB0 Go to UniProtKB: P0ACB0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ACB0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA primase | 148 | Escherichia coli K-12 | Mutation(s): 1 Gene Names: dnaG, dnaP, parB, b3066, JW3038 EC: 2.7.7.101 | 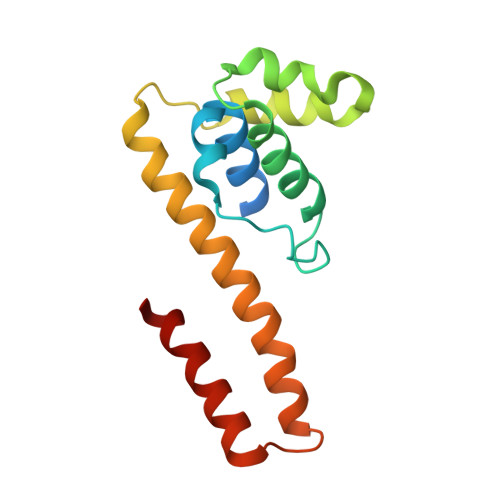 | |
UniProt | |||||
Find proteins for P0ABS5 (Escherichia coli (strain K12)) Explore P0ABS5 Go to UniProtKB: P0ABS5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ABS5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3') | J [auth M] | 20 | Escherichia coli | 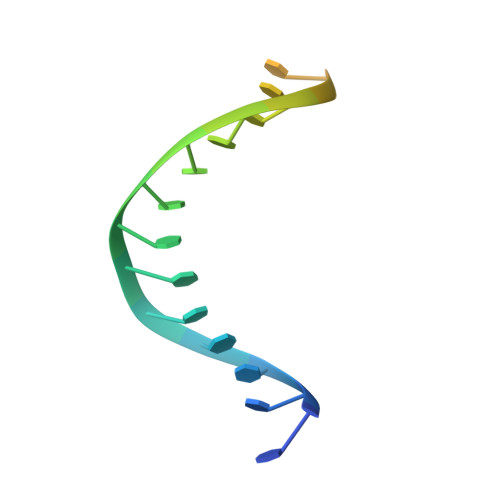 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | K [auth B], N [auth C], Q [auth D], T [auth E], W [auth F] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| ALF (Subject of Investigation/LOI) Query on ALF | L [auth B], O [auth C], R [auth D], U [auth E], X [auth F] | TETRAFLUOROALUMINATE ION Al F4 UYOMQIYKOOHAMK-UHFFFAOYSA-J |  | ||
| MG (Subject of Investigation/LOI) Query on MG | M [auth B], P [auth C], S [auth D], V [auth E], Y [auth F] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.19 |
| RECONSTRUCTION | RELION | 3.07 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Australian Research Council (ARC) | Australia | DP210100365 |