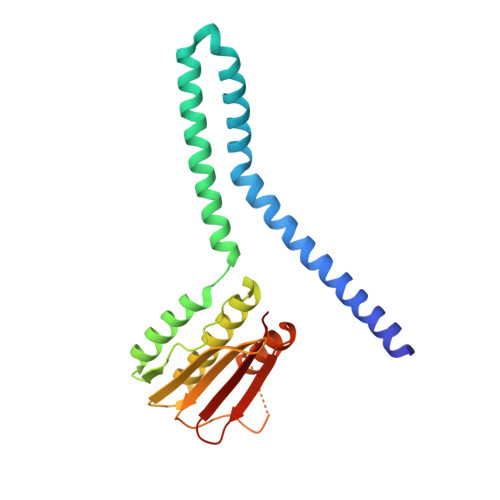
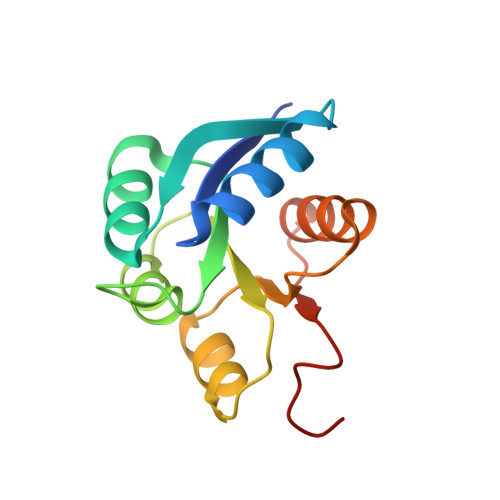
An allosteric switch ensures efficient unidirectional information transmission by the histidine kinase DesK from Bacillus subtilis.
Lima, S., Blanco, J., Olivieri, F., Imelio, J.A., Nieves, M., Carrion, F., Alvarez, B., Buschiazzo, A., Marti, M.A., Trajtenberg, F.(2023) Sci Signal 16: eabo7588-eabo7588
- PubMed: 36693130
- DOI: https://doi.org/10.1126/scisignal.abo7588
- Primary Citation of Related Structures:
7SSI, 7SSJ - PubMed Abstract:
Phosphorylation carries chemical information in biological systems. In two-component systems (TCSs), the sensor histidine kinase and the response regulator are connected through phosphoryl transfer reactions that may be uni- or bidirectional. Directionality enables the construction of complex regulatory networks that optimize signal propagation and ensure the forward flow of information. We combined x-ray crystallography, hybrid quantum mechanics/molecular mechanics (QM/MM) simulations, and systems-integrative kinetic modeling approaches to study phosphoryl flow through the Bacillus subtilis thermosensing TCS DesK-DesR. The allosteric regulation of the histidine kinase DesK was critical to avoid back transfer of phosphoryl groups and futile phosphorylation-dephosphorylation cycles by isolating phosphatase, autokinase, and phosphotransferase activities. Interactions between the kinase's ATP-binding domain and the regulator's receiver domain placed the regulator in two distinct positions in the phosphotransferase and phosphatase complexes, thereby determining whether a key glutamine residue in DesK was properly situated to assist in the dephosphorylation reaction. Moreover, an energetically unfavorable phosphotransferase conformation when DesK was not phosphorylated minimized reverse phosphoryl transfer. DesR dimerization and a dissociative phosphoryl transfer reaction also enforced the direction of phosphoryl flow. Shorter or longer distances between the phosphoryl acceptor and donor residues shifted the phosphoryl transfer equilibrium by modulating the stabilizing effect of the Mg 2+ cofactor. These mechanisms control the directionality of signal transmission and show how structure-encoded allostery stores and transmits information in signaling systems.
- Laboratory of Molecular and Structural Microbiology, Institut Pasteur de Montevideo, Montevideo, Uruguay.
Organizational Affiliation: