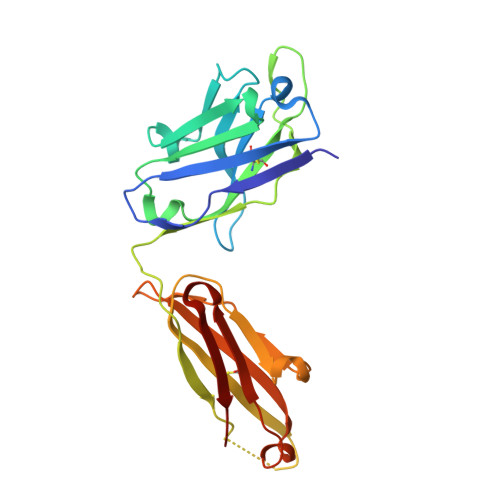
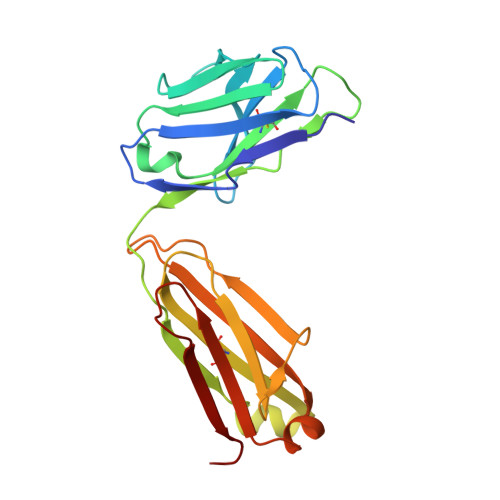
A single, improbable B cell receptor mutation confers potent neutralization against cytomegalovirus.
Jenks, J.A., Amin, S., Sponholtz, M.R., Kumar, A., Wrapp, D., Venkatayogi, S., Tu, J.J., Karthigeyan, K., Valencia, S.M., Connors, M., Harnois, M.J., Hora, B., Rochat, E., McLellan, J.S., Wiehe, K., Permar, S.R.(2023) PLoS Pathog 19: e1011107-e1011107
- PubMed: 36662906
- DOI: https://doi.org/10.1371/journal.ppat.1011107
- Primary Citation of Related Structures:
7SSC - PubMed Abstract:
Cytomegalovirus (CMV) is a leading cause of infant hearing loss and neurodevelopmental delay, but there are no clinically licensed vaccines to prevent infection, in part due to challenges eliciting neutralizing antibodies. One of the most well-studied targets for CMV vaccines is the viral fusogen glycoprotein B (gB), which is required for viral entry into host cells. Within gB, antigenic domain 2 site 1 (AD-2S1) is a target of potently neutralizing antibodies, but gB-based candidate vaccines have yet to elicit robust responses against this region. We mapped the genealogy of B cells encoding potently neutralizing anti-gB AD-2S1 antibodies from their inferred unmutated common ancestor (UCA) and characterized the binding and function of early lineage ancestors. Surprisingly, we found that a single amino acid heavy chain mutation A33N, which was an improbable mutation rarely generated by somatic hypermutation machinery, conferred broad CMV neutralization to the non-neutralizing UCA antibody. Structural studies revealed that this mutation mediated key contacts with the gB AD-2S1 epitope. Collectively, these results provide insight into potently neutralizing gB-directed antibody evolution in a single donor and lay a foundation for using this B cell-lineage directed approach for the design of next-generation CMV vaccines.
- Duke Human Vaccine Institute, Duke University Medical Center, Durham, North Carolina, United States of America.
Organizational Affiliation: