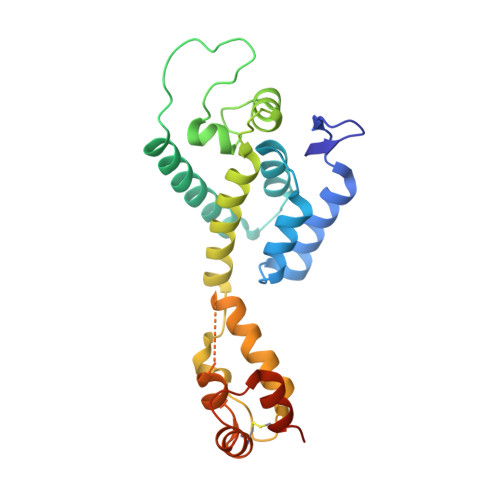
Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide
Adu-Ampratwum, D., Haney, R., Bester, S.M., Annamalai, A.S., Wei, G., Fuchs, J.R., Kvaratskhelia, M.To be published.