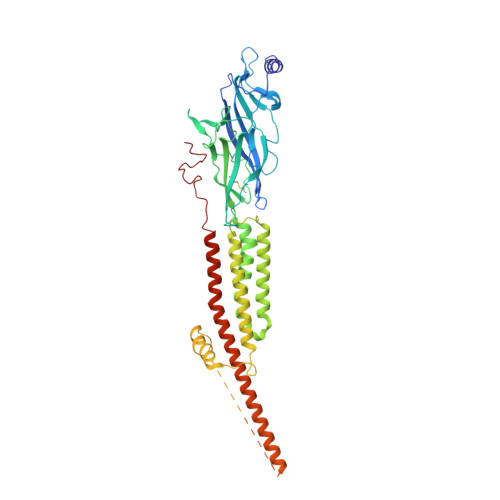
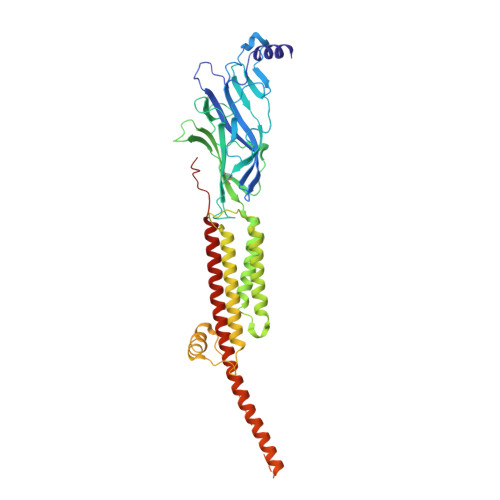
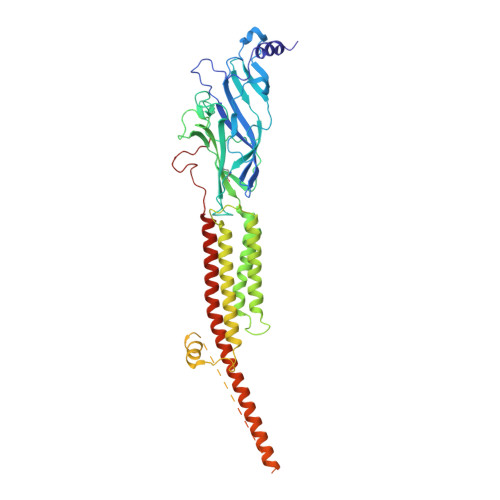
Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Rahman, M.M., Basta, T., Teng, J., Lee, M., Worrell, B.T., Stowell, M.H.B., Hibbs, R.E.(2022) Nat Struct Mol Biol 29: 386-394
- PubMed: 35301478
- DOI: https://doi.org/10.1038/s41594-022-00737-3
- Primary Citation of Related Structures:
7SMM, 7SMQ, 7SMR, 7SMS, 7SMT - PubMed Abstract:
Binding of the neurotransmitter acetylcholine to its receptors on muscle fibers depolarizes the membrane and thereby triggers muscle contraction. We sought to understand at the level of three-dimensional structure how agonists and antagonists alter nicotinic acetylcholine receptor conformation. We used the muscle-type receptor from the Torpedo ray to first define the structure of the receptor in a resting, activatable state. We then determined the receptor structure bound to the agonist carbachol, which stabilizes an asymmetric, closed channel desensitized state. We find conformational changes in a peripheral membrane helix are tied to recovery from desensitization. To probe mechanisms of antagonism, we obtained receptor structures with the active component of curare, a poison arrow toxin and precursor to modern muscle relaxants. d-Tubocurarine stabilizes the receptor in a desensitized-like state in the presence and absence of agonist. These findings define the transitions between resting and desensitized states and reveal divergent means by which antagonists block channel activity of the muscle-type nicotinic receptor.
- Department of Neuroscience, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: