Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Warren, G.M., Meir, A., Wang, J., Patel, D.J., Greene, E.C., Shuman, S.(2022) Nucleic Acids Res 50: 952-961
- PubMed: 34967418
- DOI: https://doi.org/10.1093/nar/gkab1270
- Primary Citation of Related Structures:
7SJR - PubMed Abstract:
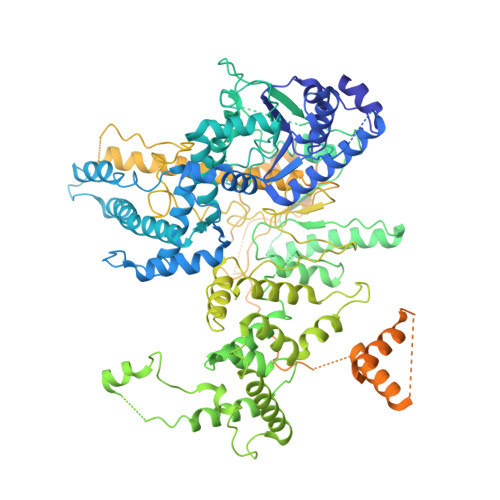
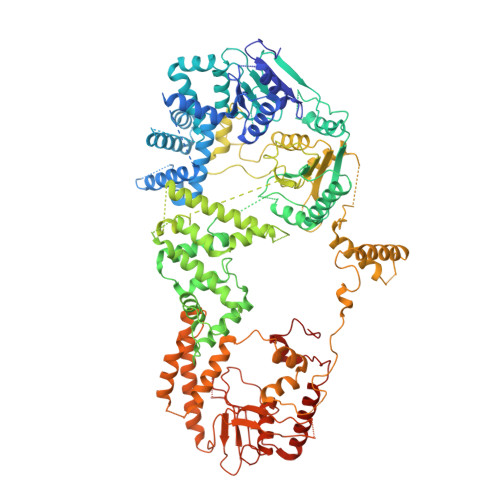
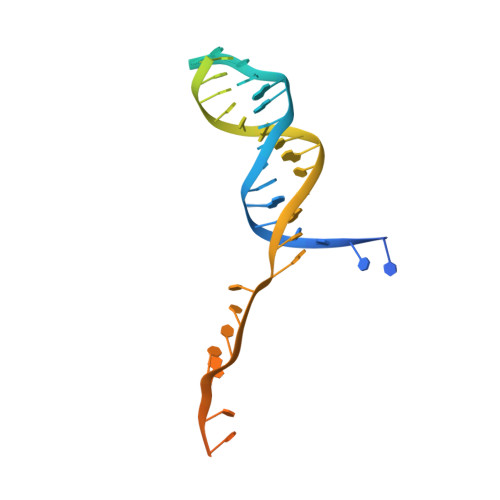
Mycobacterial AdnAB is a heterodimeric helicase-nuclease that initiates homologous recombination by resecting DNA double-strand breaks. The AdnB subunit hydrolyzes ATP to drive single-nucleotide steps of 3'-to-5' translocation of AdnAB on the tracking DNA strand via a ratchet-like mechanism. Trp325 in AdnB motif III, which intercalates into the tracking strand and makes a π stack on a nucleobase 5' of a flipped-out nucleoside, is the putative ratchet pawl without which ATP hydrolysis is mechanically futile. Here, we report that AdnAB mutants wherein Trp325 was replaced with phenylalanine, tyrosine, histidine, leucine, or alanine retained activity in ssDNA-dependent ATP hydrolysis but displayed a gradient of effects on DSB resection. The resection velocities of Phe325 and Tyr325 mutants were 90% and 85% of the wild-type AdnAB velocity. His325 slowed resection rate to 3% of wild-type and Leu325 and Ala325 abolished DNA resection. A cryo-EM structure of the DNA-bound Ala325 mutant revealed that the AdnB motif III peptide was disordered and the erstwhile flipped out tracking strand nucleobase reverted to a continuous base-stacked arrangement with its neighbors. We conclude that π stacking of Trp325 on a DNA nucleobase triggers and stabilizes the flipped-out conformation of the neighboring nucleoside that underlies formation of a ratchet pawl.
- Molecular Biology Program, Sloan Kettering Institute, New York, NY 10065, USA.
Organizational Affiliation: