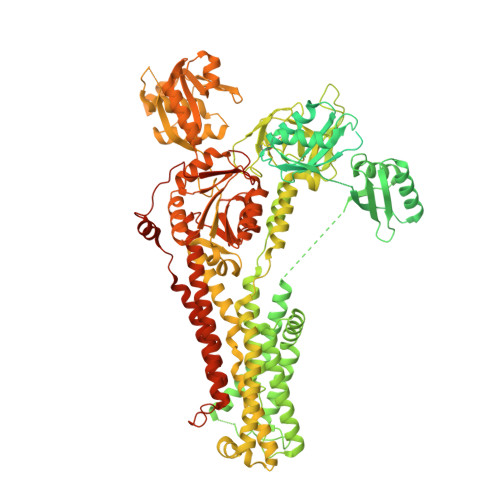
Structure of the Wilson disease copper transporter ATP7B.
Bitter, R.M., Oh, S., Deng, Z., Rahman, S., Hite, R.K., Yuan, P.(2022) Sci Adv 8: eabl5508-eabl5508
- PubMed: 35245129
- DOI: https://doi.org/10.1126/sciadv.abl5508
- Primary Citation of Related Structures:
7SI3, 7SI6, 7SI7 - PubMed Abstract:
ATP7A and ATP7B, two homologous copper-transporting P1B-type ATPases, play crucial roles in cellular copper homeostasis, and mutations cause Menkes and Wilson diseases, respectively. ATP7A/B contains a P-type ATPase core consisting of a membrane transport domain and three cytoplasmic domains, the A, P, and N domains, and a unique amino terminus comprising six consecutive metal-binding domains. Here, we present a cryo-electron microscopy structure of frog ATP7B in a copper-free state. Interacting with both the A and P domains, the metal-binding domains are poised to exert copper-dependent regulation of ATP hydrolysis coupled to transmembrane copper transport. A ring of negatively charged residues lines the cytoplasmic copper entrance that is presumably gated by a conserved basic residue sitting at the center. Within the membrane, a network of copper-coordinating ligands delineates a stepwise copper transport pathway. This work provides the first glimpse into the structure and function of ATP7 proteins and facilitates understanding of disease mechanisms and development of rational therapies.
- Department of Cell Biology and Physiology, Washington University School of Medicine, St. Louis, MO 63110, USA.
Organizational Affiliation: