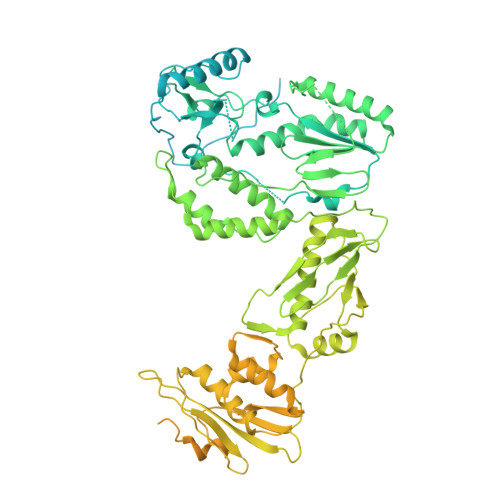
Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Harrison, J.J.E.K., Passos, D.O., Bruhn, J.F., Bauman, J.D., Tuberty, L., DeStefano, J.J., Ruiz, F.X., Lyumkis, D., Arnold, E.(2022) Sci Adv 8: eabn9874-eabn9874
- PubMed: 35857464
- DOI: https://doi.org/10.1126/sciadv.abn9874
- Primary Citation of Related Structures:
7SEP, 7SJX - PubMed Abstract:
Key proteins of retroviruses and other RNA viruses are translated and subsequently processed from polyprotein precursors by the viral protease (PR). Processing of the HIV Gag-Pol polyprotein yields the HIV structural proteins and enzymes. Structures of the mature enzymes PR, reverse transcriptase (RT), and integrase (IN) aided understanding of catalysis and design of antiretrovirals, but knowledge of the Pol precursor architecture and function before PR cleavage is limited. We developed a system to produce stable HIV-1 Pol and determined its cryo-electron microscopy structure. RT in Pol has a similar arrangement to the mature RT heterodimer, and its dimerization may draw together two PR monomers to activate proteolytic processing. HIV-1 thus may leverage the dimerization interfaces in Pol to regulate assembly and maturation of polyprotein precursors.
- Center for Advanced Biotechnology and Medicine (CABM), Piscataway, NJ, USA.
Organizational Affiliation: