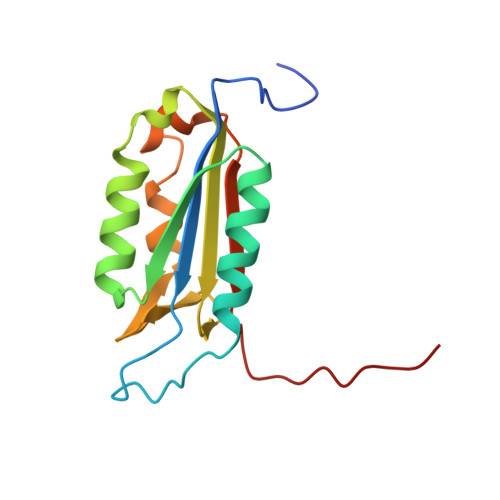
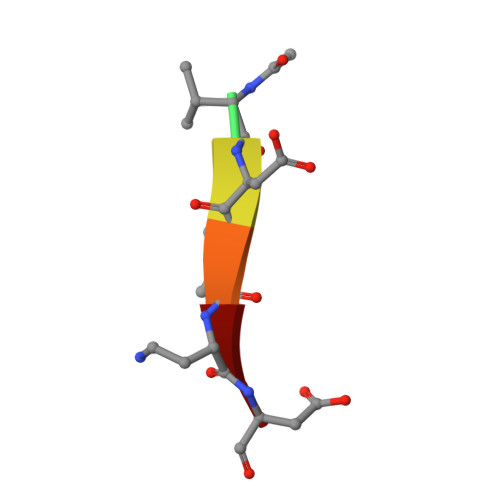
Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Bresinsky, M., Strasser, J.M., Vallaster, B., Liu, P., McCue, W.M., Fuller, J., Hubmann, A., Singh, G., Nelson, K.M., Cuellar, M.E., Wilmot, C.M., Finzel, B.C., Ashe, K.H., Walters, M.A., Pockes, S.(2022) Acs Pharmacol Transl Sci 5: 20-40
- PubMed: 35059567
- DOI: https://doi.org/10.1021/acsptsci.1c00251
- Primary Citation of Related Structures:
7RN7, 7RN8, 7RN9, 7RNB, 7RND, 7RNE, 7RNF, 7SEO - PubMed Abstract:
Alzheimer's disease (AD) was first described by Alois Alzheimer over 100 years ago, but there is still no overarching theory that can explain its cause in detail. There are also no effective therapies to treat either the cause or the associated symptoms of this devastating disease. A potential approach to better understand the pathogenesis of AD could be the development of selective caspase-2 (Casp2) probes, as we have shown that a Casp2-mediated cleavage product of tau (Δtau314) reversibly impairs cognitive and synaptic function in animal models of tauopathies. In this article, we map out the Casp2 binding site through the preparation and assay of a series of 35 pentapeptide inhibitors with the goal of gaining selectivity against caspase-3 (Casp3). We also employed computational docking methods to understand the key interactions in the binding pocket of Casp2 and the differences predicted for binding at Casp3. Moreover, we crystallographically characterized the binding of selected pentapeptides with Casp3. Furthermore, we engineered and expressed a series of recombinant tau mutants and investigated them in an in vitro cleavage assay. These studies resulted in simple peptidic inhibitors with nanomolar affinity, for example, AcVDV(Dab)D-CHO ( 24 ) with up to 27.7-fold selectivity against Casp3. Our findings provide a good basis for the future development of selective Casp2 probes and inhibitors that can serve as pharmacological tools in planned in vivo studies and as lead compounds for the design of bioavailable and more drug-like small molecules.
- Institute of Pharmacy, University of Regensburg, Universitätsstraße 31, Regensburg 93053, Germany.
Organizational Affiliation: