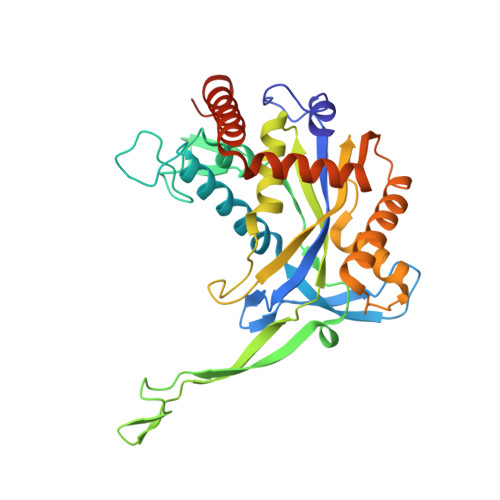
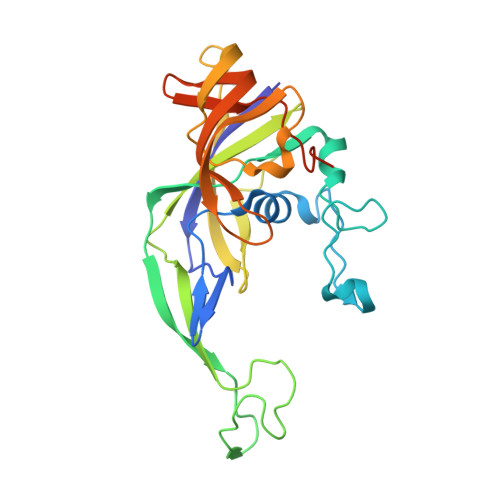
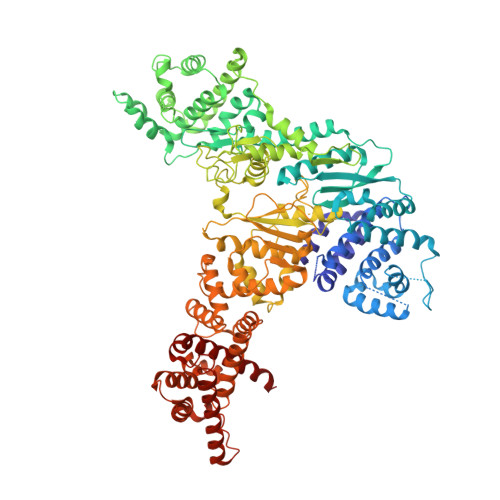
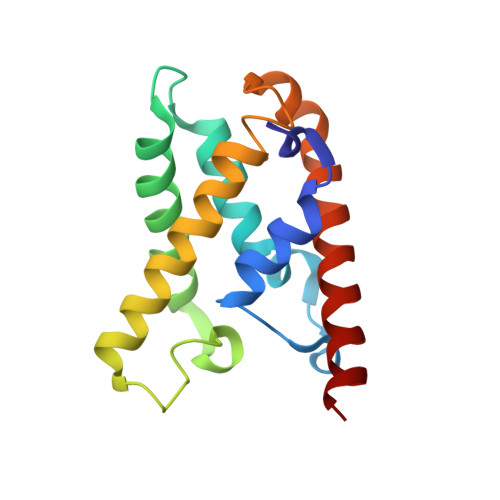
Structural rearrangements allow nucleic acid discrimination by type I-D Cascade.
Schwartz, E.A., McBride, T.M., Bravo, J.P.K., Wrapp, D., Fineran, P.C., Fagerlund, R.D., Taylor, D.W.(2022) Nat Commun 13: 2829-2829
- PubMed: 35595728
- DOI: https://doi.org/10.1038/s41467-022-30402-8
- Primary Citation of Related Structures:
7SBA, 7SBB - PubMed Abstract:
CRISPR-Cas systems are adaptive immune systems that protect prokaryotes from foreign nucleic acids, such as bacteriophages. Two of the most prevalent CRISPR-Cas systems include type I and type III. Interestingly, the type I-D interference proteins contain characteristic features of both type I and type III systems. Here, we present the structures of type I-D Cascade bound to both a double-stranded (ds)DNA and a single-stranded (ss)RNA target at 2.9 and 3.1 Å, respectively. We show that type I-D Cascade is capable of specifically binding ssRNA and reveal how PAM recognition of dsDNA targets initiates long-range structural rearrangements that likely primes Cas10d for Cas3' binding and subsequent non-target strand DNA cleavage. These structures allow us to model how binding of the anti-CRISPR protein AcrID1 likely blocks target dsDNA binding via competitive inhibition of the DNA substrate engagement with the Cas10d active site. This work elucidates the unique mechanisms used by type I-D Cascade for discrimination of single-stranded and double stranded targets. Thus, our data supports a model for the hybrid nature of this complex with features of type III and type I systems.
- Department of Molecular Biosciences, University of Texas at Austin, Austin, TX, 78712-1597, USA.
Organizational Affiliation: