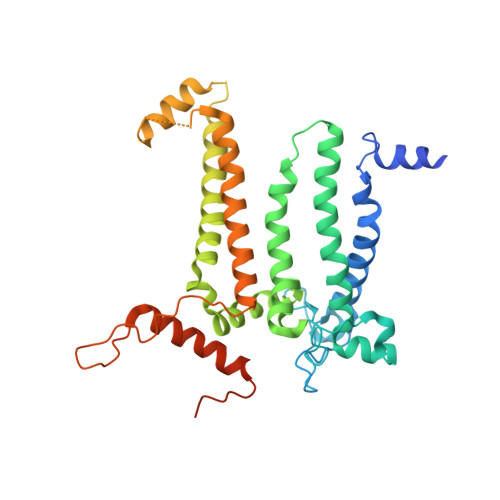
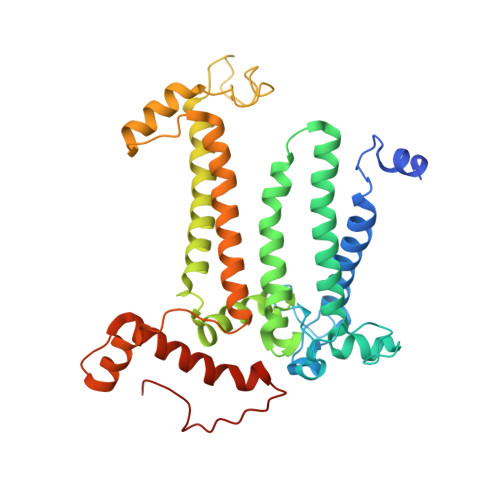
Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light reveals the functions of chlorophylls d and f.
Gisriel, C.J., Shen, G., Ho, M.Y., Kurashov, V., Flesher, D.A., Wang, J., Armstrong, W.H., Golbeck, J.H., Gunner, M.R., Vinyard, D.J., Debus, R.J., Brudvig, G.W., Bryant, D.A.(2021) J Biological Chem 298: 101424-101424
- PubMed: 34801554
- DOI: https://doi.org/10.1016/j.jbc.2021.101424
- Primary Citation of Related Structures:
7SA3 - PubMed Abstract:
Far-red light (FRL) photoacclimation in cyanobacteria provides a selective growth advantage for some terrestrial cyanobacteria by expanding the range of photosynthetically active radiation to include far-red/near-infrared light (700-800 nm). During this photoacclimation process, photosystem II (PSII), the water:plastoquinone photooxidoreductase involved in oxygenic photosynthesis, is modified. The resulting FRL-PSII is comprised of FRL-specific core subunits and binds chlorophyll (Chl) d and Chl f molecules in place of several of the Chl a molecules found when cells are grown in visible light. These new Chls effectively lower the energy canonically thought to define the "red limit" for light required to drive photochemical catalysis of water oxidation. Changes to the architecture of FRL-PSII were previously unknown, and the positions of Chl d and Chl f molecules had only been proposed from indirect evidence. Here, we describe the 2.25 Å resolution cryo-EM structure of a monomeric FRL-PSII core complex from Synechococcus sp. PCC 7335 cells that were acclimated to FRL. We identify one Chl d molecule in the Chl D1 position of the electron transfer chain and four Chl f molecules in the core antenna. We also make observations that enhance our understanding of PSII biogenesis, especially on the acceptor side of the complex where a bicarbonate molecule is replaced by a glutamate side chain in the absence of the assembly factor Psb28. In conclusion, these results provide a structural basis for the lower energy limit required to drive water oxidation, which is the gateway for most solar energy utilization on earth.
- Department of Chemistry, Yale University, New Haven, Connecticut, USA.
Organizational Affiliation: