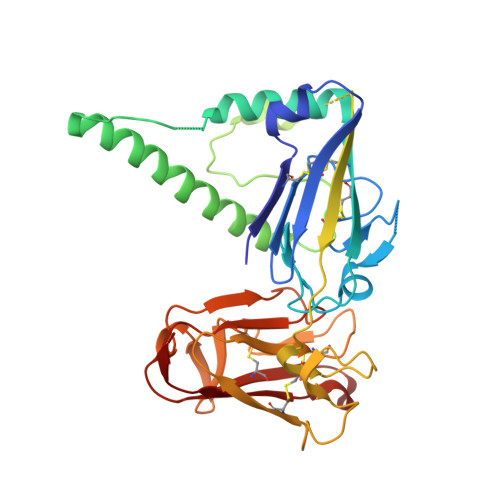
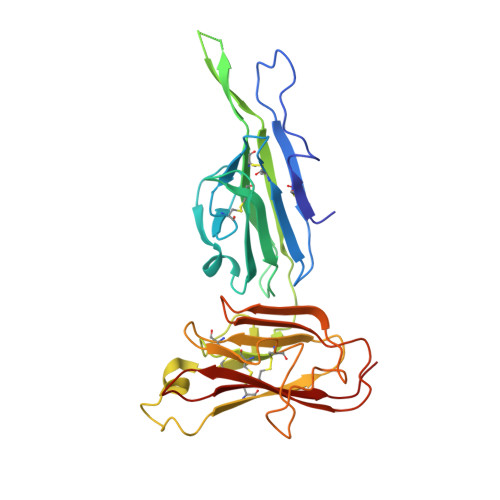
Structure of the Pf12 and Pf41 heterodimeric complex of Plasmodium falciparum 6-cysteine proteins.
Dietrich, M.H., Chan, L.J., Adair, A., Boulet, C., O'Neill, M.T., Tan, L.L., Keremane, S., Mok, Y.F., Lo, A.W., Gilson, P., Tham, W.H.(2022) FEMS Microbes 3: xtac005-xtac005
- PubMed: 35308105
- DOI: https://doi.org/10.1093/femsmc/xtac005
- Primary Citation of Related Structures:
7S7Q, 7S7R - PubMed Abstract:
During the different stages of the Plasmodium life cycle, surface-associated proteins establish key interactions with the host and play critical roles in parasite survival. The 6-cysteine (6-cys) protein family is one of the most abundant surface antigens and expressed throughout the Plasmodium falciparum life cycle. This protein family is conserved across Plasmodium species and plays critical roles in parasite transmission, evasion of the host immune response and host cell invasion. Several 6-cys proteins are present on the parasite surface as hetero-complexes but it is not known how two 6-cys proteins interact together. Here, we present a crystal structure of Pf12 bound to Pf41 at 2.85 Å resolution, two P. falciparum proteins usually found on the parasite surface of late schizonts and merozoites. Our structure revealed two critical interfaces required for complex formation with important implications on how different 6-cysteine proteins may interact with each other. Using structure-function analyses, we identified important residues for Pf12-Pf41 complex formation. In addition, we generated 16 nanobodies against Pf12 and Pf41 and showed that several Pf12-specific nanobodies inhibit Pf12-Pf41 complex formation. Using X-ray crystallography, we were able to describe the structural mechanism of an inhibitory nanobody in blocking Pf12-Pf41 complex formation. Future studies using these inhibitory nanobodies will be useful to determine the functional role of these two 6-cys proteins in malaria parasites.
- Infectious Diseases and Immune Defence Division, The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria3052, Australia.
Organizational Affiliation: