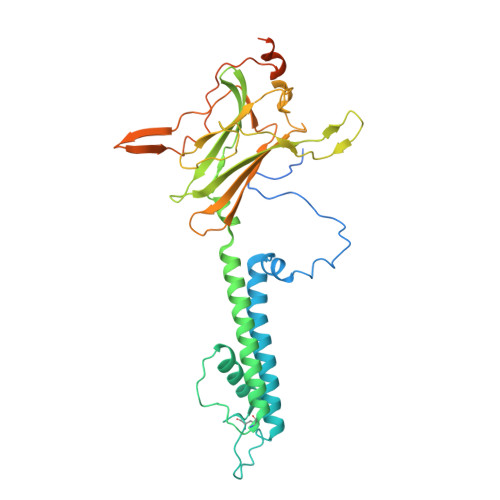
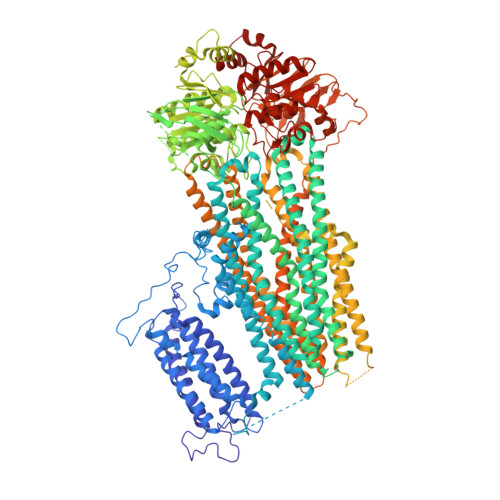
Molecular structure of an open human K ATP channel.
Zhao, C., MacKinnon, R.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34815345
- DOI: https://doi.org/10.1073/pnas.2112267118
- Primary Citation of Related Structures:
7S5T, 7S5V, 7S5X, 7S5Y, 7S5Z, 7S60, 7S61 - PubMed Abstract:
K ATP channels are metabolic sensors that translate intracellular ATP/ADP balance into membrane excitability. The molecular composition of K ATP includes an inward-rectifier potassium channel (Kir) and an ABC transporter-like sulfonylurea receptor (SUR). Although structures of K ATP have been determined in many conformations, in all cases, the pore in Kir is closed. Here, we describe human pancreatic K ATP (hK ATP ) structures with an open pore at 3.1- to 4.0-Å resolution using single-particle cryo-electron microscopy (cryo-EM). Pore opening is associated with coordinated structural changes within the ATP-binding site and the channel gate in Kir. Conformational changes in SUR are also observed, resulting in an area reduction of contact surfaces between SUR and Kir. We also observe that pancreatic hK ATP exhibits the unique (among inward-rectifier channels) property of PIP 2 -independent opening, which appears to be correlated with a docked cytoplasmic domain in the absence of PIP 2 .
- HHMI, The Rockefeller University, New York, NY 10065.
Organizational Affiliation: