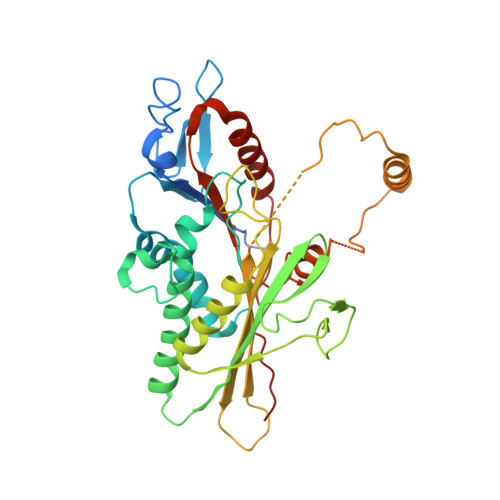
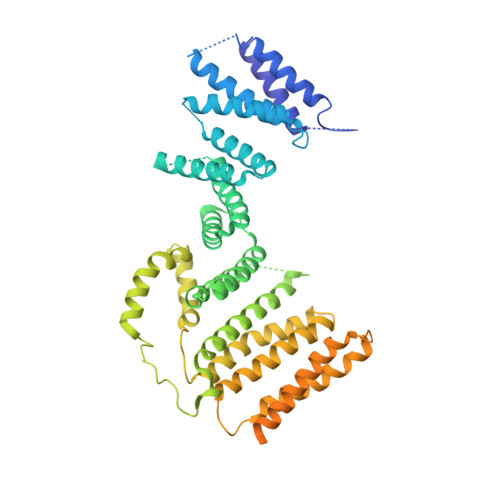
Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Solon, A.L., Tan, Z., Schutt, K.L., Jepsen, L., Haynes, S.E., Nesvizhskii, A.I., Sept, D., Stumpff, J., Ohi, R., Cianfrocco, M.A.(2021) Sci Adv 7: eabj9812-eabj9812
- PubMed: 34797717
- DOI: https://doi.org/10.1126/sciadv.abj9812
- Primary Citation of Related Structures:
7RSI, 7RSQ, 7RYP, 7RYQ - PubMed Abstract:
Kinesins are regulated in space and time to ensure activation only in the presence of cargo. Kinesin-binding protein (KIFBP), which is mutated in Goldberg-Shprintzen syndrome, binds to and inhibits the catalytic motor heads of 8 of 45 kinesin superfamily members, but the mechanism remains poorly defined. Here, we used cryo–electron microscopy and cross-linking mass spectrometry to determine high-resolution structures of KIFBP alone and in complex with two mitotic kinesins, revealing structural remodeling of kinesin by KIFBP. We find that KIFBP remodels kinesin motors and blocks microtubule binding (i) via allosteric changes to kinesin and (ii) by sterically blocking access to the microtubule. We identified two regions of KIFBP necessary for kinesin binding and cellular regulation during mitosis. Together, this work further elucidates the molecular mechanism of KIFBP-mediated kinesin inhibition and supports a model in which structural rearrangement of kinesin motor domains by KIFBP abrogates motor protein activity.
- Department of Cell and Developmental Biology, University of Michigan, Ann Arbor, MI, USA.
Organizational Affiliation: