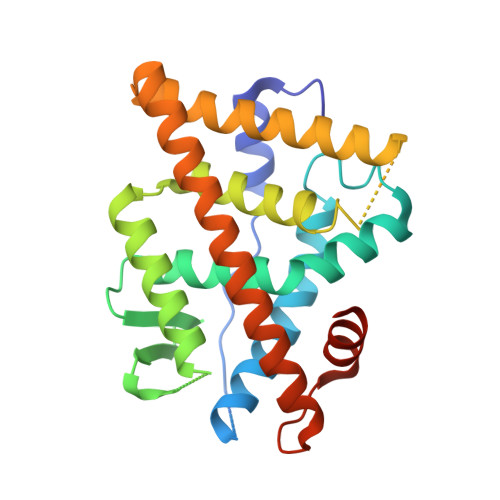
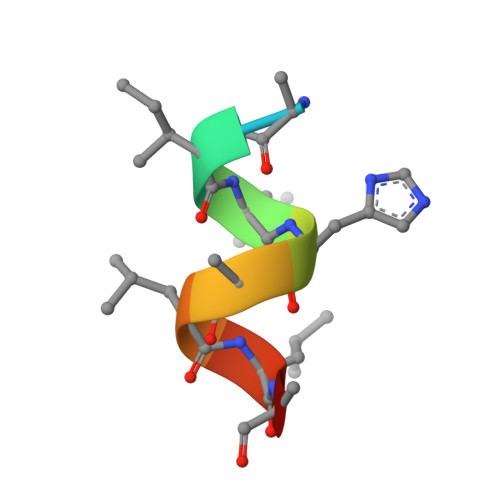
Structural Determinants of the Binding and Activation of Estrogen Receptor alpha by Phenolic Thieno[2,3-d]pyrimidines
Reddy Sammeta, V., Anderson, B.M., Norris, J.D., Torrice, C.D., Joiner, C., Liu, S., Li, H., Popov, K.I., Fanning, S.W., McDonnell, D.P., Willson, T.M.(2023) Helvetica Chim Acta 106