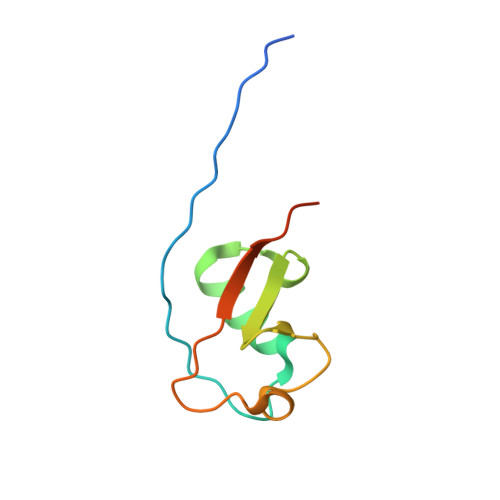
Panel of Engineered Ubiquitin Variants Targeting the Family of Human Ubiquitin Interacting Motifs.
Veggiani, G., Yates, B.P., Martyn, G.D., Manczyk, N., Singer, A.U., Kurinov, I., Sicheri, F., Sidhu, S.S.(2022) ACS Chem Biol 17: 941-956
- PubMed: 35385646
- DOI: https://doi.org/10.1021/acschembio.2c00089
- Primary Citation of Related Structures:
7RMA - PubMed Abstract:
Ubiquitin (Ub)-binding domains embedded in intracellular proteins act as readers of the complex Ub code and contribute to regulation of numerous eukaryotic processes. Ub-interacting motifs (UIMs) are short α-helical modular recognition elements whose role in controlling proteostasis and signal transduction has been poorly investigated. Moreover, impaired or aberrant activity of UIM-containing proteins has been implicated in numerous diseases, but targeting modular recognition elements in proteins remains a major challenge. To overcome this limitation, we developed Ub variants (UbVs) that bind to 42 UIMs in the human proteome with high affinity and specificity. Structural analysis of a UbV:UIM complex revealed the molecular determinants of enhanced affinity and specificity. Furthermore, we showed that a UbV targeting a UIM in the cancer-associated Ub-specific protease 28 potently inhibited catalytic activity. Our work demonstrates the versatility of UbVs to target short α-helical Ub receptors with high affinity and specificity. Moreover, the UbVs provide a toolkit to investigate the role of UIMs in regulating and transducing Ub signals and establish a general strategy for the systematic development of probes for Ub-binding domains.
- Donnelly Centre for Cellular and Biomolecular Research, Banting and Best Department of Medical Research, University of Toronto, 160 College Street, Toronto, Ontario M5S3E1, Canada.
Organizational Affiliation: