Crystal structure of human N-myristoyltransferase 1 fragment (residues 109-496) bound to diacylated human Arf6 octapeptide and Coenzyme A
Fenwick, M.K., Lin, H.To be published.
Experimental Data Snapshot
Starting Model: other
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycylpeptide N-tetradecanoyltransferase 1 | 391 | Homo sapiens | Mutation(s): 0 Gene Names: NMT1, NMT EC: 2.3.1.97 (PDB Primary Data), 2.3.1 (UniProt) | 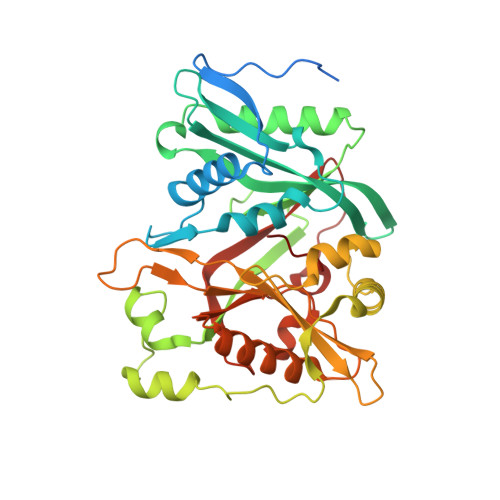 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P30419 (Homo sapiens) Explore P30419 Go to UniProtKB: P30419 | |||||
PHAROS: P30419 GTEx: ENSG00000136448 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30419 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ADP-ribosylation factor 6 | 8 | Homo sapiens | Mutation(s): 0 Gene Names: ARF6 EC: 3.6.5.2 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62330 (Homo sapiens) Explore P62330 Go to UniProtKB: P62330 | |||||
PHAROS: P62330 GTEx: ENSG00000165527 | |||||
Entity Groups | |||||
| UniProt Group | P62330 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 4PS (Subject of Investigation/LOI) Query on 4PS | C [auth A] | 4'-diphospho pantetheine C11 H24 N2 O10 P2 S UQURMDBHCKDEJS-VIFPVBQESA-N |  | ||
| MYR (Subject of Investigation/LOI) Query on MYR | D [auth B] | MYRISTIC ACID C14 H28 O2 TUNFSRHWOTWDNC-UHFFFAOYSA-N |  | ||
| 6NA (Subject of Investigation/LOI) Query on 6NA | E [auth B] | HEXANOIC ACID C6 H12 O2 FUZZWVXGSFPDMH-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 47.984 | α = 90 |
| b = 76.046 | β = 90 |
| c = 106.342 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| PHENIX | phasing |
| Aimless | data scaling |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Disease (NIH/NIDDK) | United States | DK107868-04 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM124165-01 |
| National Institutes of Health/Office of the Director | S10OD021527 | |
| Department of Energy (DOE, United States) | United States | DE-AC02-06CH11357 |