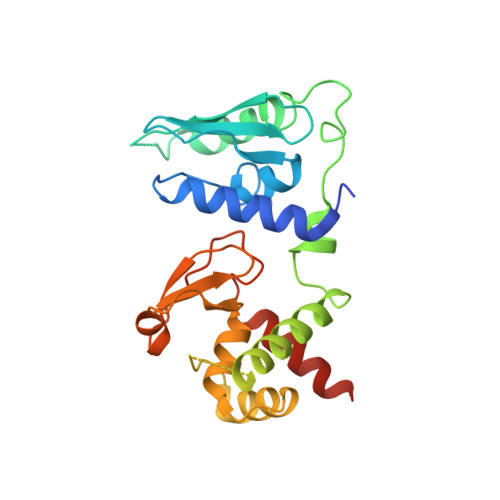
Structure of the pre-mRNA leakage 39-kDa protein reveals a single domain of integrated zf-C3HC and Rsm1 modules.
Hashimoto, H., Ramirez, D.H., Lautier, O., Pawlak, N., Blobel, G., Palancade, B., Debler, E.W.(2022) Sci Rep 12: 17691-17691
- PubMed: 36271106
- DOI: https://doi.org/10.1038/s41598-022-22183-3
- Primary Citation of Related Structures:
7RDN - PubMed Abstract:
In Saccharomyces cerevisiae, the pre-mRNA leakage 39-kDa protein (ScPml39) was reported to retain unspliced pre-mRNA prior to export through nuclear pore complexes (NPCs). Pml39 homologs outside the Saccharomycetaceae family are currently unknown, and mechanistic insight into Pml39 function is lacking. Here we determined the crystal structure of ScPml39 at 2.5 Å resolution to facilitate the discovery of orthologs beyond Saccharomycetaceae, e.g. in Schizosaccharomyces pombe or human. The crystal structure revealed integrated zf-C3HC and Rsm1 modules, which are tightly associated through a hydrophobic interface to form a single domain. Both zf-C3HC and Rsm1 modules belong to the Zn-containing BIR (Baculovirus IAP repeat)-like super family, with key residues of the canonical BIR domain being conserved. Features unique to the Pml39 modules refer to the spacing between the Zn-coordinating residues, giving rise to a substantially tilted helix αC in the zf-C3HC and Rsm1 modules, and an extra helix αAB' in the Rsm1 module. Conservation of key residues responsible for its distinct features identifies S. pombe Rsm1 and Homo sapiens NIPA/ZC3HC1 as structural orthologs of ScPml39. Based on the recent functional characterization of NIPA/ZC3HC1 as a scaffold protein that stabilizes the nuclear basket of the NPC, our data suggest an analogous function of ScPml39 in S. cerevisiae.
- Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, PA, 19107, USA.
Organizational Affiliation: