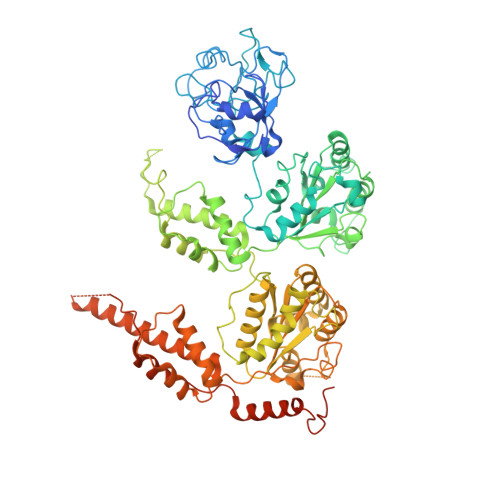
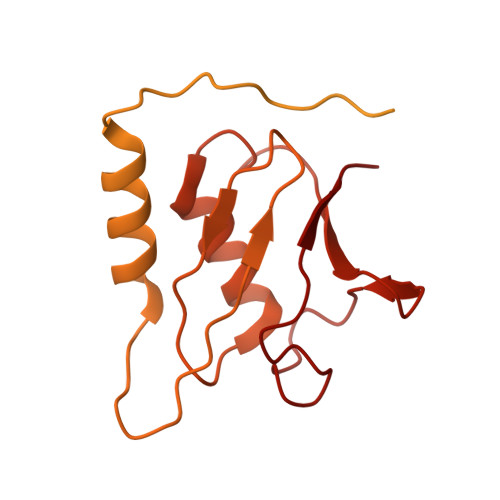
Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Nandi, P., Li, S., Columbres, R.C.A., Wang, F., Williams, D.R., Poh, Y.P., Chou, T.F., Chiu, P.L.(2021) Int J Mol Sci 22
- PubMed: 34360842
- DOI: https://doi.org/10.3390/ijms22158079
- Primary Citation of Related Structures:
7L5W, 7L5X, 7R7S, 7R7T, 7R7U - PubMed Abstract:
IBMPFD/ALS is a genetic disorder caused by a single amino acid mutation on the p97 ATPase, promoting ATPase activity and cofactor dysregulation. The disease mechanism underlying p97 ATPase malfunction remains unclear. To understand how the mutation alters the ATPase regulation, we assembled a full-length p97 R155H with its p47 cofactor and first visualized their structures using single-particle cryo-EM. More than one-third of the population was the dodecameric form. Nucleotide presence dissociates the dodecamer into two hexamers for its highly elevated function. The N-domains of the p97 R155H mutant all show up configurations in ADP- or ATP γ S-bound states. Our functional and structural analyses showed that the p47 binding is likely to impact the p97 R155H ATPase activities via changing the conformations of arginine fingers. These functional and structural analyses underline the ATPase dysregulation with the miscommunication between the functional modules of the p97 R155H .
- Biodesign Center for Applied Structural Discovery, School of Molecular Sciences, Arizona State University, Tempe, AZ 85287, USA.
Organizational Affiliation: