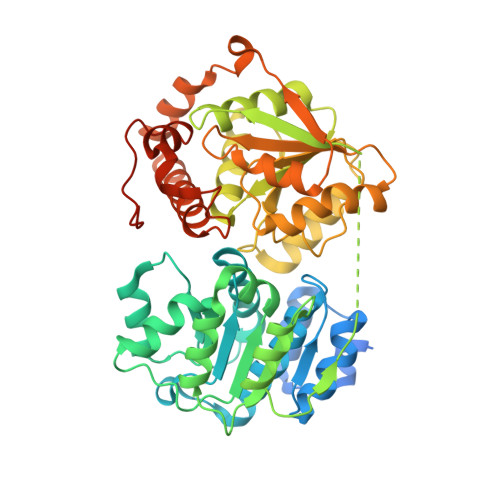
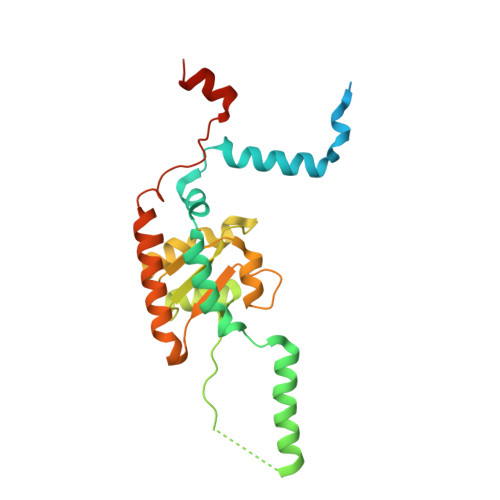
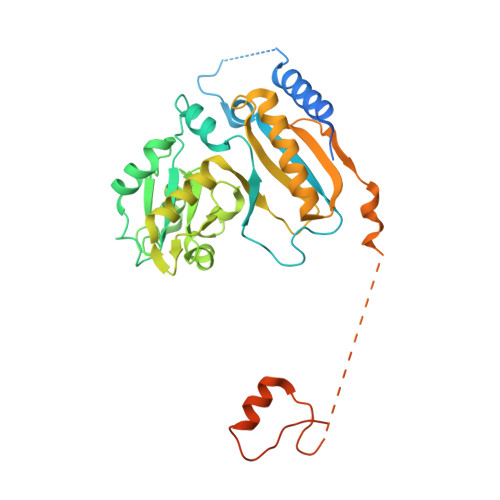
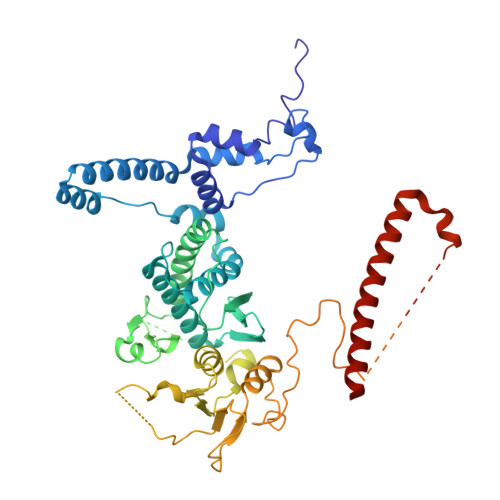
Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Cruz, V.E., Sekulski, K., Peddada, N., Sailer, C., Balasubramanian, S., Weirich, C.S., Stengel, F., Erzberger, J.P.(2022) Nat Struct Mol Biol 29: 1228-1238
- PubMed: 36482249
- DOI: https://doi.org/10.1038/s41594-022-00874-9
- Primary Citation of Related Structures:
7NAC, 7NAD, 7NAF, 7R6K, 7R6Q, 7R72, 7R7A, 7R7C, 7U0H - PubMed Abstract:
DEAD-box ATPases are ubiquitous enzymes essential in all aspects of RNA biology. However, the limited in vitro catalytic activities described for these enzymes are at odds with their complex cellular roles, most notably in driving large-scale RNA remodeling steps during the assembly of ribonucleoproteins (RNPs). We describe cryo-EM structures of 60S ribosomal biogenesis intermediates that reveal how context-specific RNA unwinding by the DEAD-box ATPase Spb4 results in extensive, sequence-specific remodeling of rRNA secondary structure. Multiple cis and trans interactions stabilize Spb4 in a post-catalytic, high-energy intermediate that drives the organization of the three-way junction at the base of rRNA domain IV. This mechanism explains how limited strand separation by DEAD-box ATPases is leveraged to provide non-equilibrium directionality and ensure efficient and accurate RNP assembly.
- Department of Biophysics, UT Southwestern Medical Center - ND10.124B, Dallas, TX, USA.
Organizational Affiliation: