CARM1 Transition State Mimics
Marechal, N., Cura, V., Troffer-Charlier, N., Bonnefond, L., Cavarelli, J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone-arginine methyltransferase CARM1 | 378 | Mus musculus | Mutation(s): 0 Gene Names: Carm1, Prmt4 EC: 2.1.1.319 | 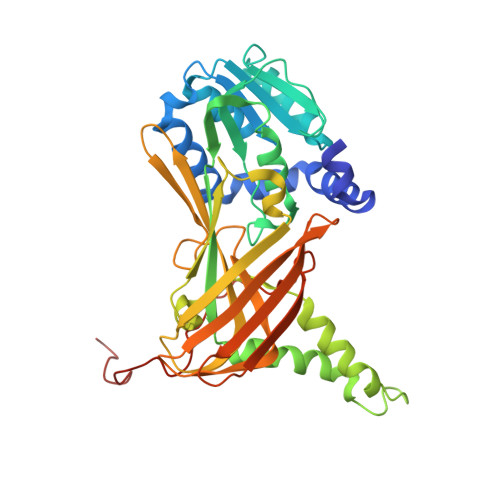 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9WVG6 (Mus musculus) Explore Q9WVG6 Go to UniProtKB: Q9WVG6 | |||||
IMPC: MGI:1913208 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9WVG6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SER-THR-GLY-GLY-LYS-ALA-PRO-URU-LYS-GLN-LEU-ALA-THR-LYS-ALA-ALA | E [auth L], F [auth M] | 16 | Mus musculus | Mutation(s): 0 | 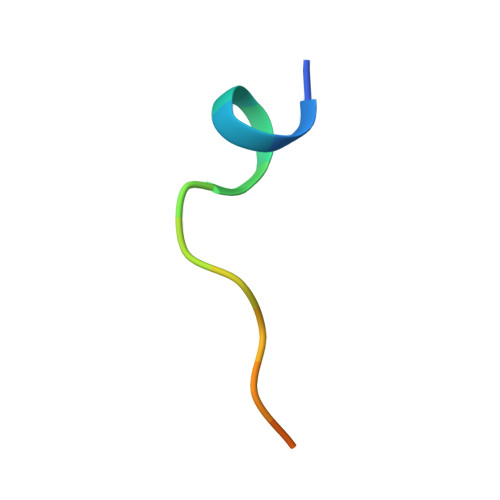 |
UniProt | |||||
Find proteins for P68433 (Mus musculus) Explore P68433 Go to UniProtKB: P68433 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68433 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SAH Query on SAH | J [auth B], M [auth D] | S-ADENOSYL-L-HOMOCYSTEINE C14 H20 N6 O5 S ZJUKTBDSGOFHSH-WFMPWKQPSA-N |  | ||
| QVR (Subject of Investigation/LOI) Query on QVR | O [auth L], P [auth M] | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol C12 H15 N5 O3 UYHMWDPUDJRZGB-JVINVVEESA-N |  | ||
| MLI Query on MLI | I [auth B], N [auth L] | MALONATE ION C3 H2 O4 OFOBLEOULBTSOW-UHFFFAOYSA-L |  | ||
| EDO Query on EDO | G [auth B], H [auth B], K [auth D], L [auth D] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 75.183 | α = 90 |
| b = 99.204 | β = 90 |
| c = 208.25 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| XDS | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |