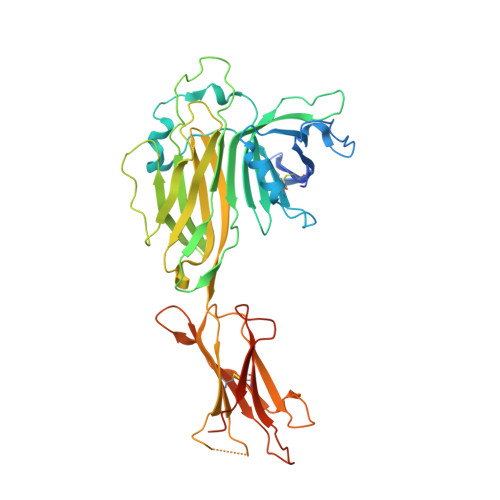
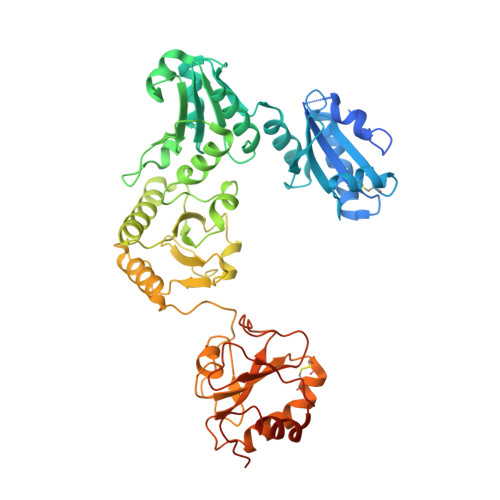
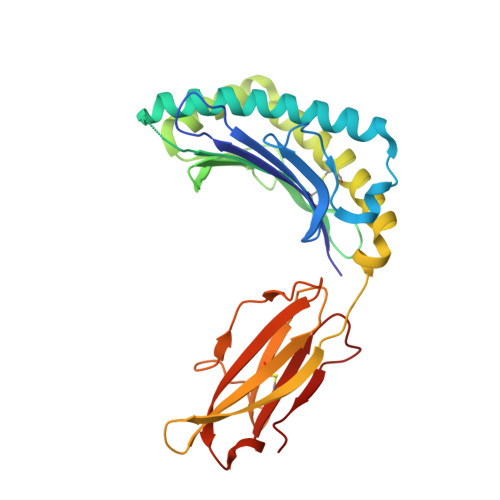
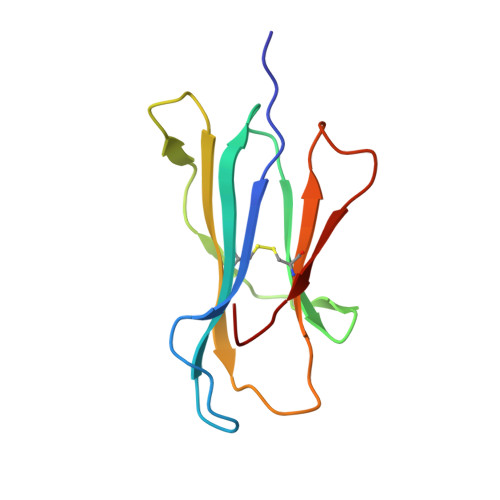
Structure of an MHC I-tapasin-ERp57 editing complex defines chaperone promiscuity.
Muller, I.K., Winter, C., Thomas, C., Spaapen, R.M., Trowitzsch, S., Tampe, R.(2022) Nat Commun 13: 5383-5383
- PubMed: 36104323
- DOI: https://doi.org/10.1038/s41467-022-32841-9
- Primary Citation of Related Structures:
7QNG - PubMed Abstract:
Adaptive immunity depends on cell surface presentation of antigenic peptides by major histocompatibility complex class I (MHC I) molecules and on stringent ER quality control in the secretory pathway. The chaperone tapasin in conjunction with the oxidoreductase ERp57 is crucial for MHC I assembly and for shaping the epitope repertoire for high immunogenicity. However, how the tapasin-ERp57 complex engages MHC I clients has not yet been determined at atomic detail. Here, we present the 2.7-Å crystal structure of a tapasin-ERp57 heterodimer in complex with peptide-receptive MHC I. Our study unveils molecular details of client recognition by the multichaperone complex and highlights elements indispensable for peptide proofreading. The structure of this transient ER quality control complex provides the mechanistic basis for the selector function of tapasin and showcases how the numerous MHC I allomorphs are chaperoned during peptide loading and editing.
- Institute of Biochemistry, Biocenter, Goethe University Frankfurt, Frankfurt/Main, Germany.
Organizational Affiliation: