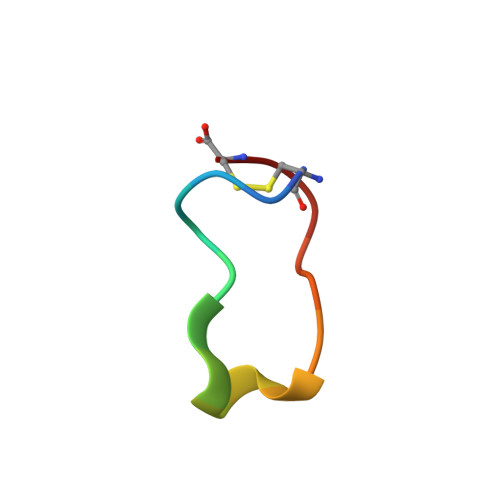
Conformational ensemble of the TNF-derived peptide solnatide in solution.
Martin-Malpartida, P., Arrastia-Casado, S., Farrera-Sinfreu, J., Lucas, R., Fischer, H., Fischer, B., Eaton, D.C., Tzotzos, S., Macias, M.J.(2022) Comput Struct Biotechnol J 20: 2082-2090
- PubMed: 35601958
- DOI: https://doi.org/10.1016/j.csbj.2022.04.031
- Primary Citation of Related Structures:
7QLF - PubMed Abstract:
Tumor necrosis factor (TNF) is a homotrimer that has two spatially distinct binding regions, three lectin-like domains (LLD) at the TIP of the protein and three basolaterally located receptor-binding sites, the latter of which are responsible for the inflammatory and cell death-inducing properties of the cytokine. Solnatide (a.k.a. TIP peptide, AP301) is a 17-mer cyclic peptide that mimics the LLD of human TNF which activates the amiloride-sensitive epithelial sodium channel (ENaC) and, as such, recapitulates the capacity of TNF to enhance alveolar fluid clearance, as demonstrated in numerous preclinical studies. TNF and solnatide interact with glycoproteins and these interactions are necessary for their trypanolytic and ENaC-activating activities. In view of the crucial role of ENaC in lung liquid clearance, solnatide is currently being evaluated as a novel therapeutic agent to treat pulmonary edema in patients with moderate-to-severe acute respiratory distress syndrome (ARDS), as well as severe COVID-19 patients with ARDS. To facilitate the description of the functional properties of solnatide in detail, as well as to further target-docking studies, we have analyzed its folding properties by NMR. In solution, solnatide populates a set of conformations characterized by a small hydrophobic core and two electrostatically charged poles. Using the structural information determined here and also that available for the ENaC protein, we propose a model to describe solnatide interaction with the C-terminal domain of the ENaCα subunit. This model may serve to guide future experiments to validate specific interactions with ENaCα and the design of new solnatide analogs with unexplored functionalities.
- Institute for Research in Biomedicine, The Barcelona Institute of Science and Technology, Baldiri Reixac, 10, Barcelona 08028, Spain.
Organizational Affiliation: