Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization
Morichaud, Z., Trapani, S., Vishwakarma, R.K., Chaloin, L., Lionne, C., Lai-Kee-Him, J., Bron, P., Brodolin, K.(2023) Nat Commun 14: 484
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2023) Nat Commun 14: 484
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit alpha | 347 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: rpoA, Rv3457c, MTCY13E12.10c EC: 2.7.7.6 | 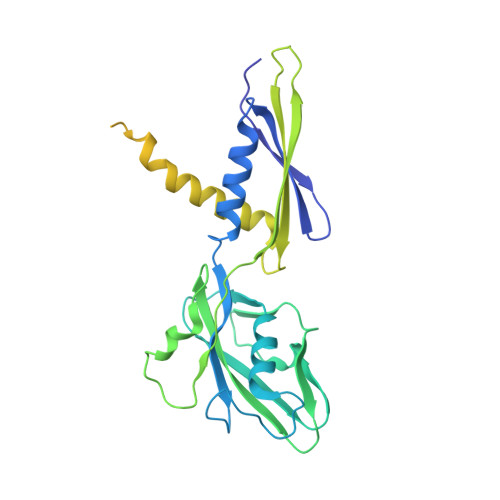 | |
UniProt | |||||
Find proteins for P9WGZ1 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WGZ1 Go to UniProtKB: P9WGZ1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WGZ1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit beta | 1,174 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: rpoB, Rv0667, MTCI376.08c EC: 2.7.7.6 | 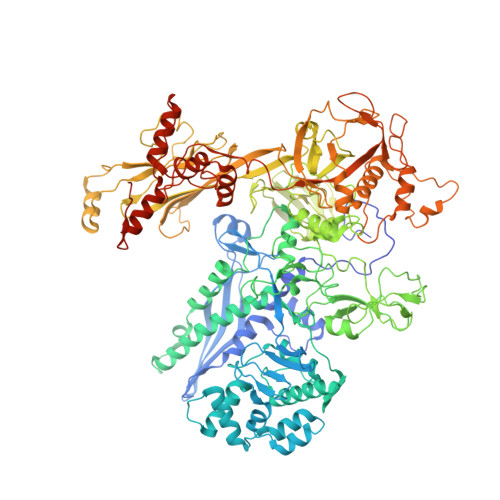 | |
UniProt | |||||
Find proteins for P9WGY9 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WGY9 Go to UniProtKB: P9WGY9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WGY9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit beta' | 1,319 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: rpoC, Rv0668, MTCI376.07c EC: 2.7.7.6 | 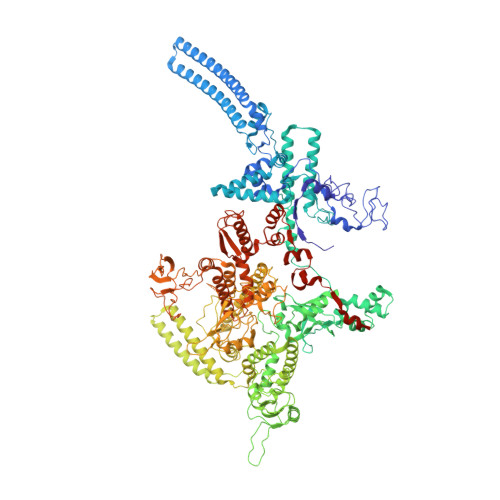 | |
UniProt | |||||
Find proteins for P9WGY7 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WGY7 Go to UniProtKB: P9WGY7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WGY7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit omega | 110 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: rpoZ, Rv1390, MTCY21B4.07 EC: 2.7.7.6 | 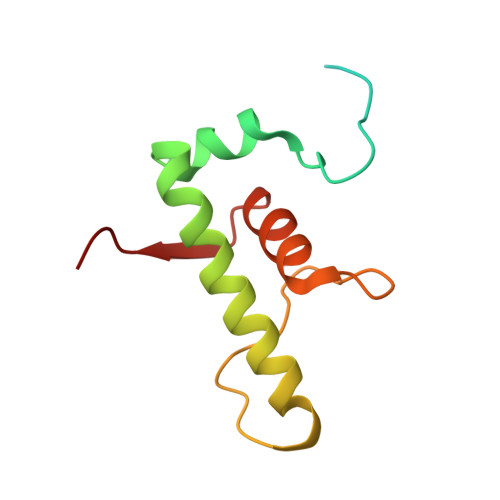 | |
UniProt | |||||
Find proteins for P9WGY5 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WGY5 Go to UniProtKB: P9WGY5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WGY5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA polymerase sigma factor SigB | 343 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: sigB, mysB, Rv2710 |  | |
UniProt | |||||
Find proteins for P9WGI5 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WGI5 Go to UniProtKB: P9WGI5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WGI5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN (Subject of Investigation/LOI) Query on ZN | AB [auth J] CB [auth P] DB [auth P] FB [auth W] GB [auth W] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | BB [auth J] EB [auth P] HB [auth W] KB [auth CA] NB [auth IA] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC | 3.1 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Agence Nationale de la Recherche (ANR) | France | ANR-16-CE11-0025 |