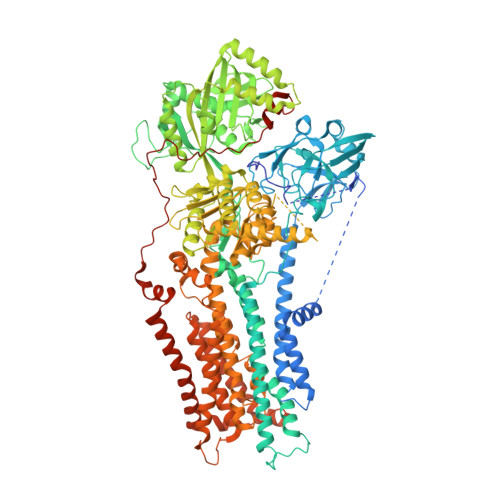
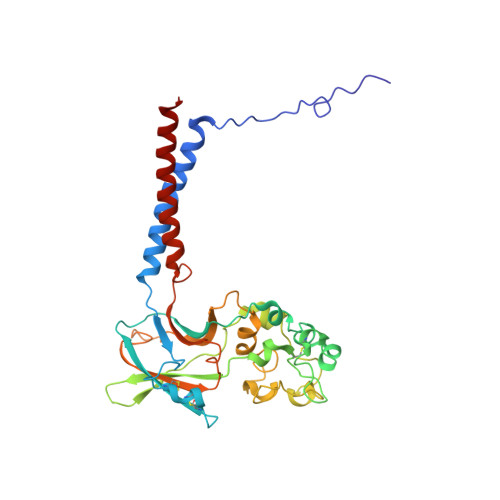
Autoinhibition and regulation by phosphoinositides of ATP8B1, a human lipid flippase associated with intrahepatic cholestatic disorders.
Dieudonne, T., Herrera, S.A., Laursen, M.J., Lejeune, M., Stock, C., Slimani, K., Jaxel, C., Lyons, J.A., Montigny, C., Pomorski, T.G., Nissen, P., Lenoir, G.(2022) Elife 11
- PubMed: 35416773
- DOI: https://doi.org/10.7554/eLife.75272
- Primary Citation of Related Structures:
7PY4 - PubMed Abstract:
P4-ATPases flip lipids from the exoplasmic to the cytosolic leaflet, thus maintaining lipid asymmetry in eukaryotic cell membranes. Mutations in several human P4-ATPase genes are associated with severe diseases, for example in ATP8B1 causing progressive familial intrahepatic cholestasis, a rare inherited disorder progressing toward liver failure. ATP8B1 forms a binary complex with CDC50A and displays a broad specificity to glycerophospholipids, but regulatory mechanisms are unknown. Here, we report functional studies and the cryo-EM structure of the human lipid flippase ATP8B1-CDC50A at 3.1 Å resolution. We find that ATP8B1 is autoinhibited by its N- and C-terminal tails, which form extensive interactions with the catalytic sites and flexible domain interfaces. Consistently, ATP hydrolysis is unleashed by truncation of the C-terminus, but also requires phosphoinositides, most markedly phosphatidylinositol-3,4,5-phosphate (PI(3,4,5)P 3 ), and removal of both N- and C-termini results in full activation. Restored inhibition of ATP8B1 truncation constructs with a synthetic peptide mimicking the C-terminal segment further suggests molecular communication between N- and C-termini in the autoinhibition and demonstrates that the regulatory mechanism can be interfered with by exogenous compounds. A recurring (G/A)(Y/F)AFS motif of the C-terminal segment suggests that this mechanism is employed widely across P4-ATPase lipid flippases in plasma membrane and endomembranes.
- Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), Gif-sur-Yvette, France.
Organizational Affiliation: