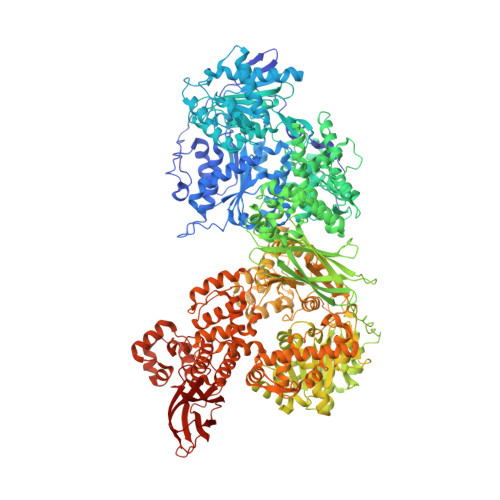
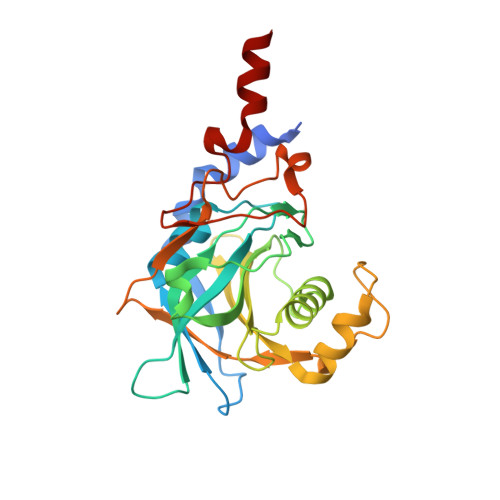
The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Bergfort, A., Hilal, T., Kuropka, B., Ilik, I.A., Weber, G., Aktas, T., Freund, C., Wahl, M.C.(2022) Nucleic Acids Res 50: 2938-2958
- PubMed: 35188580
- DOI: https://doi.org/10.1093/nar/gkac087
- Primary Citation of Related Structures:
7OS1, 7OS2, 7PX3 - PubMed Abstract:
Biogenesis of spliceosomal small nuclear ribonucleoproteins (snRNPs) and their recycling after splicing require numerous assembly/recycling factors whose modes of action are often poorly understood. The intrinsically disordered TSSC4 protein has been identified as a nuclear-localized U5 snRNP and U4/U6-U5 tri-snRNP assembly/recycling factor, but how TSSC4's intrinsic disorder supports TSSC4 functions remains unknown. Using diverse interaction assays and cryogenic electron microscopy-based structural analysis, we show that TSSC4 employs four conserved, non-contiguous regions to bind the PRPF8 Jab1/MPN domain and the SNRNP200 helicase at functionally important sites. It thereby inhibits SNRNP200 helicase activity, spatially aligns the proteins, coordinates formation of a U5 sub-module and transiently blocks premature interaction of SNRNP200 with at least three other spliceosomal factors. Guided by the structure, we designed a TSSC4 variant that lacks stable binding to the PRPF8 Jab1/MPN domain or SNRNP200 in vitro. Comparative immunoprecipitation/mass spectrometry from HEK293 nuclear extract revealed distinct interaction profiles of wild type TSSC4 and the variant deficient in PRPF8/SNRNP200 binding with snRNP proteins, other spliceosomal proteins as well as snRNP assembly/recycling factors and chaperones. Our findings elucidate molecular strategies employed by an intrinsically disordered protein to promote snRNP assembly, and suggest multiple TSSC4-dependent stages during snRNP assembly/recycling.
- Freie Universität Berlin, Institute of Chemistry and Biochemistry, Laboratory of Structural Biochemistry, Takustr. 6, D-14195 Berlin, Germany.
Organizational Affiliation: