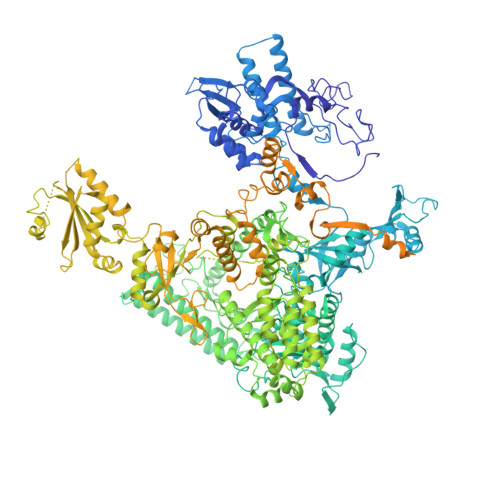
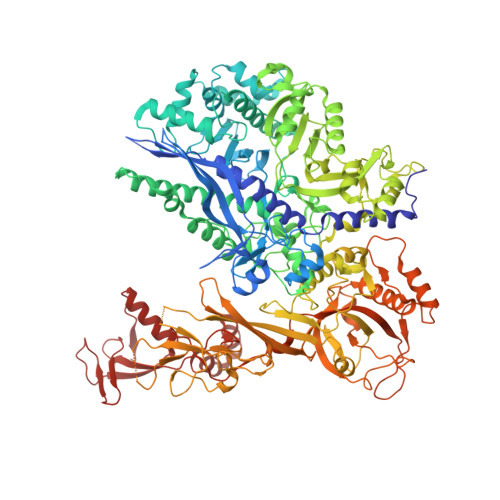
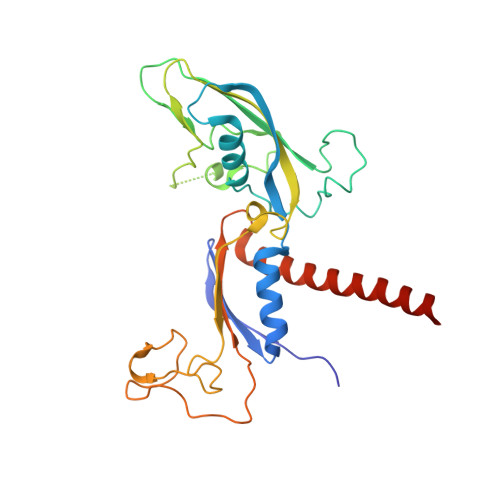
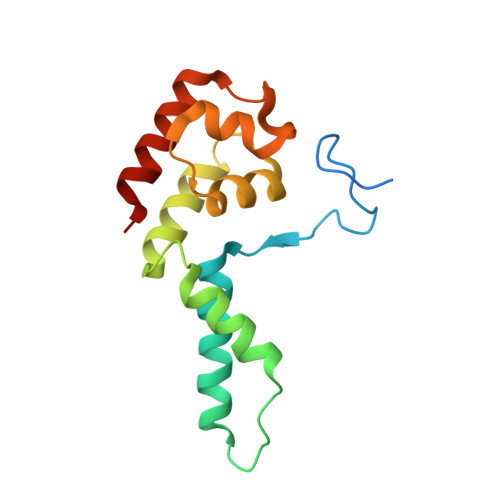
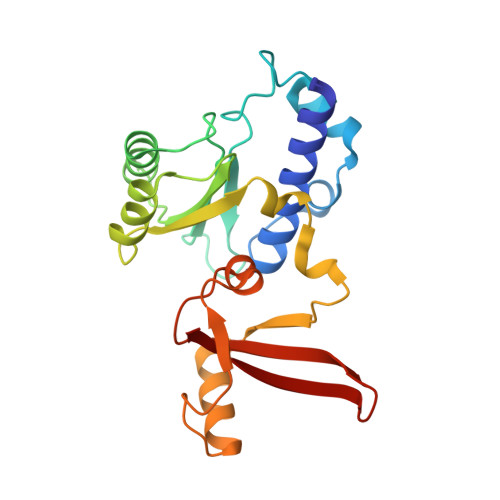
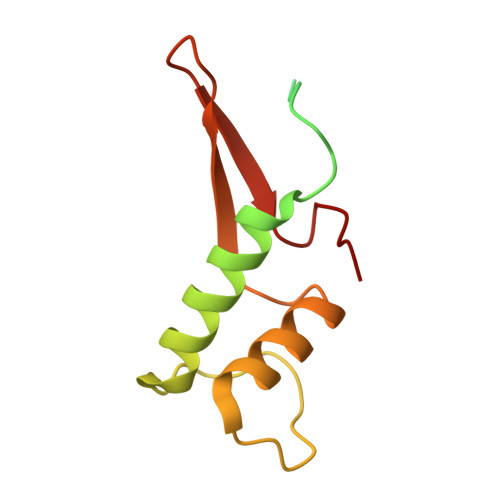
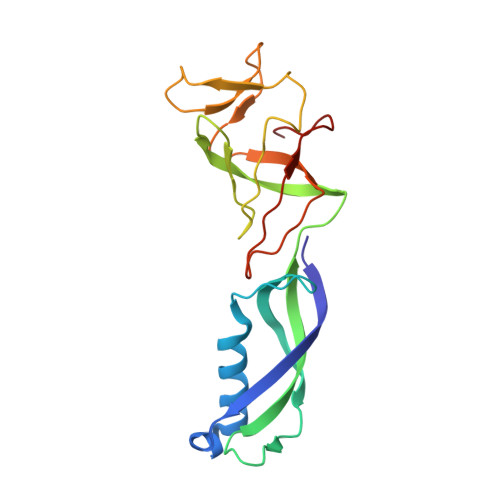
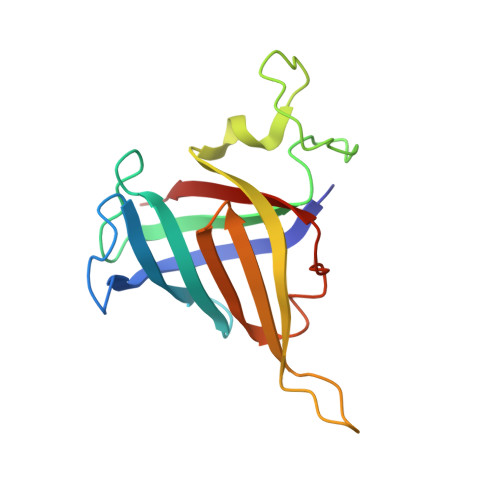
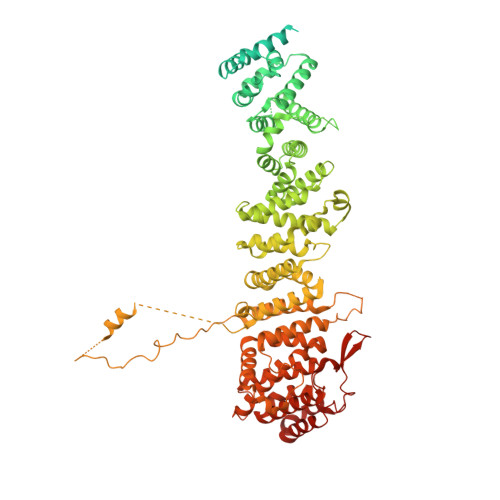
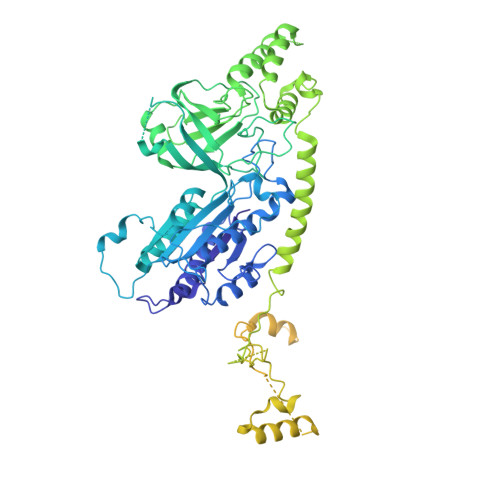
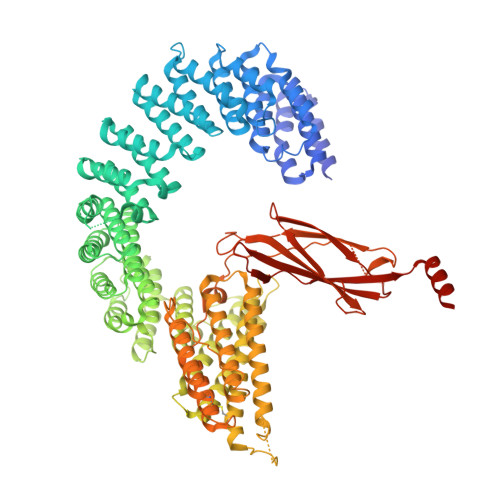
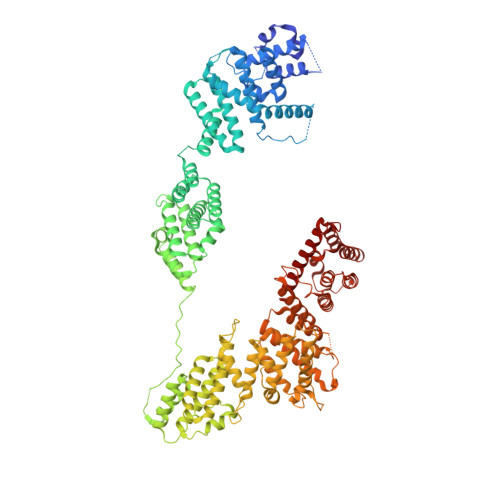
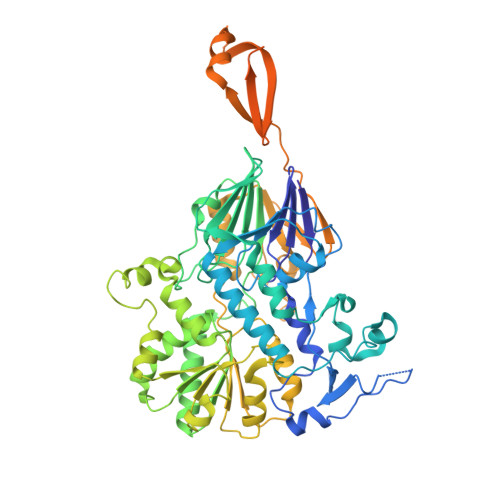
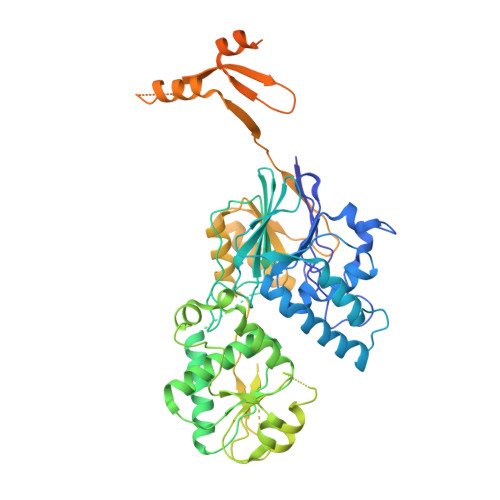
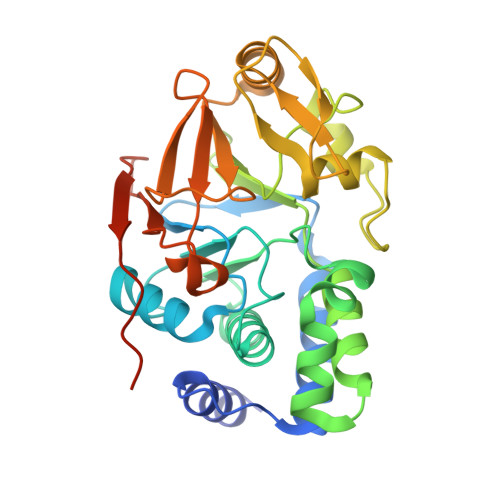
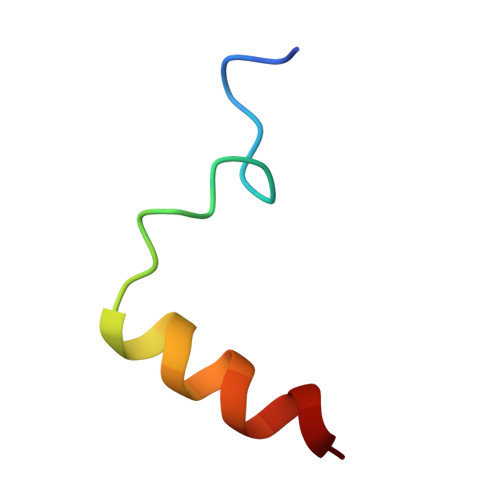
Structural basis of Integrator-mediated transcription regulation.
Fianu, I., Chen, Y., Dienemann, C., Dybkov, O., Linden, A., Urlaub, H., Cramer, P.(2021) Science 374: 883-887
- PubMed: 34762484
- DOI: https://doi.org/10.1126/science.abk0154
- Primary Citation of Related Structures:
7PKS - PubMed Abstract:
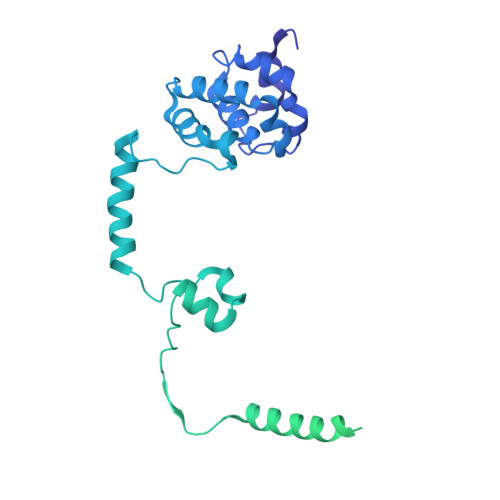
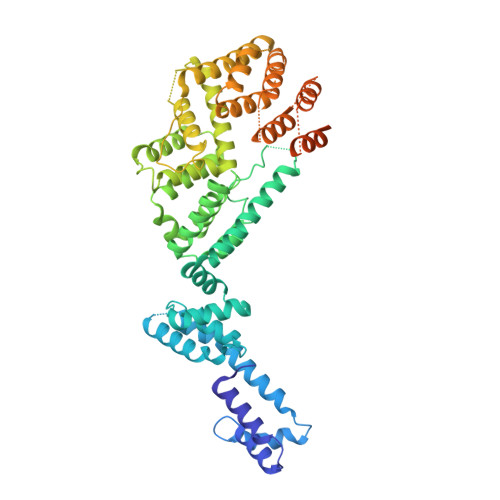
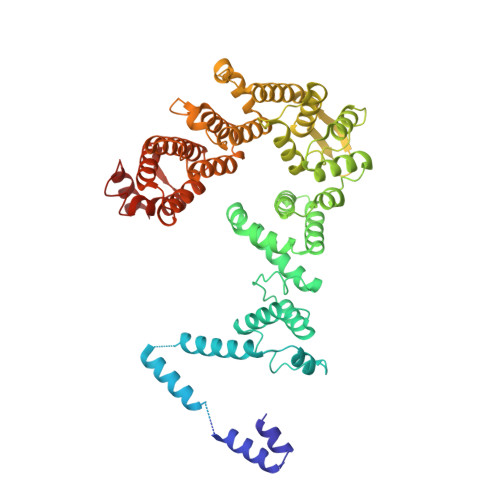
Integrator and protein phosphatase 2A (PP2A) form a complex that dephosphorylates paused RNA polymerase II (Pol II), cleaves the nascent RNA, and terminates transcription. We report the structure of the pretermination complex containing the human Integrator-PP2A complex bound to paused Pol II. Integrator binds Pol II and the pausing factors DSIF and NELF to exclude binding of the elongation factors SPT6 and PAF1 complex. Integrator also binds the C-terminal domain of Pol II and positions PP2A to counteract Pol II phosphorylation and elongation. The Integrator endonuclease docks to the RNA exit site and opens to cleave nascent RNA about 20 nucleotides from the Pol II active site. Integrator does not bind the DNA clamps formed by Pol II and DSIF, enabling release of DNA and transcription termination.
- Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany.
Organizational Affiliation: