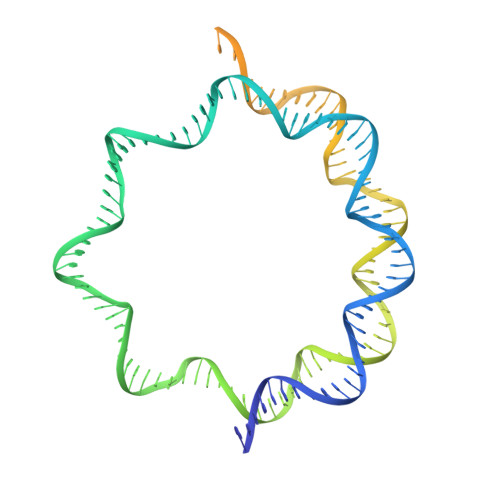
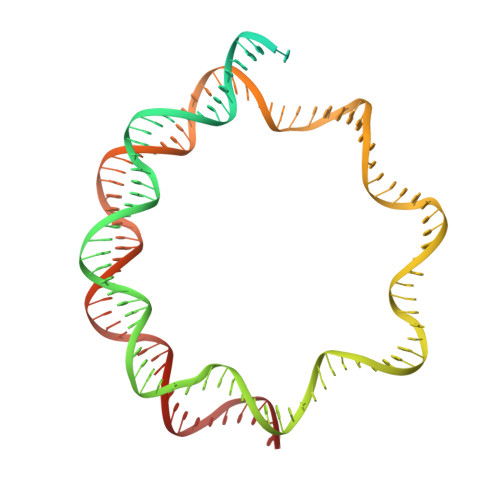
Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Yatskevich, S., Muir, K.W., Bellini, D., Zhang, Z., Yang, J., Tischer, T., Predin, M., Dendooven, T., McLaughlin, S.H., Barford, D.(2022) Science 376: 844-852
- PubMed: 35420891
- DOI: https://doi.org/10.1126/science.abn3810
- Primary Citation of Related Structures:
7PB4, 7PB8, 7PII, 7PKN, 7R5R, 7R5S, 7R5V, 7YWX, 7YYH - PubMed Abstract:
Kinetochores assemble onto specialized centromeric CENP-A (centromere protein A) nucleosomes (CENP-A Nuc ) to mediate attachments between chromosomes and the mitotic spindle. We describe cryo-electron microscopy structures of the human inner kinetochore constitutive centromere associated network (CCAN) complex bound to CENP-A Nuc reconstituted onto α-satellite DNA. CCAN forms edge-on contacts with CENP-A Nuc , and a linker DNA segment of the α-satellite repeat emerges from the fully wrapped end of the nucleosome to thread through the central CENP-LN channel that tightly grips the DNA. The CENP-TWSX histone-fold module further augments DNA binding and partially wraps the linker DNA in a manner reminiscent of canonical nucleosomes. Our study suggests that the topological entrapment of the linker DNA by CCAN provides a robust mechanism by which kinetochores withstand both pushing and pulling forces exerted by the mitotic spindle.
- MRC Laboratory of Molecular Biology, Cambridge CB2 0QH, UK.
Organizational Affiliation: