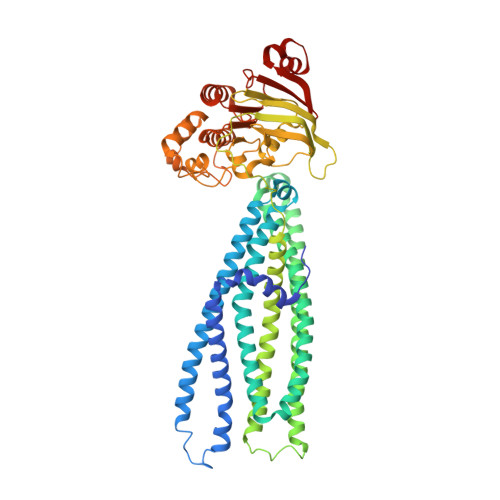
The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Galazzo, L., Meier, G., Januliene, D., Parey, K., De Vecchis, D., Striednig, B., Hilbi, H., Schafer, L.V., Kuprov, I., Moeller, A., Bordignon, E., Seeger, M.A.(2022) Sci Adv 8: eabn6845-eabn6845
- PubMed: 36223470
- DOI: https://doi.org/10.1126/sciadv.abn6845
- Primary Citation of Related Structures:
7NDF, 7PH2, 7PH3, 7PH4, 7PH7 - PubMed Abstract:
Membrane proteins are currently investigated after detergent extraction from native cellular membranes and reconstitution into artificial liposomes or nanodiscs, thereby removing them from their physiological environment. However, to truly understand the biophysical properties of membrane proteins in a physiological environment, they must be investigated within living cells. Here, we used a spin-labeled nanobody to interrogate the conformational cycle of the ABC transporter MsbA by double electron-electron resonance. Unexpectedly, the wide inward-open conformation of MsbA, commonly considered a nonphysiological state, was found to be prominently populated in Escherichia coli cells. Molecular dynamics simulations revealed that extensive lateral portal opening is essential to provide access of its large natural substrate core lipid A to the binding cavity. Our work paves the way to investigate the conformational landscape of membrane proteins in cells.
- Faculty of Chemistry and Biochemistry, Ruhr University Bochum, 44801 Bochum, Germany.
Organizational Affiliation: