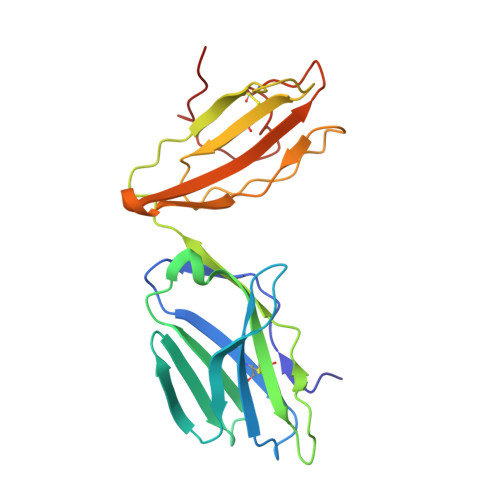
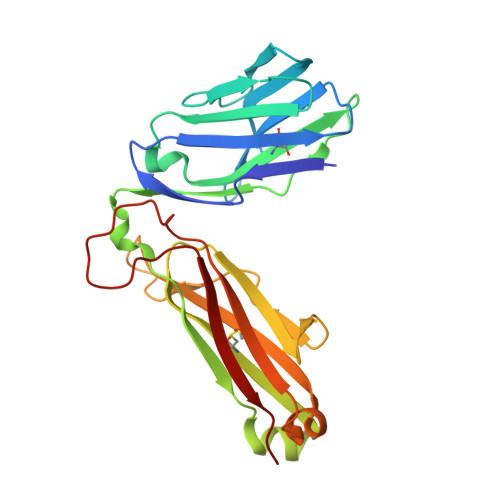
Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
Simister, P.C., Border, E.C., Vieira, J.F., Pumphrey, N.J.(2022) J Immunother Cancer 10
- PubMed: 35851311
- DOI: https://doi.org/10.1136/jitc-2022-004600
- Primary Citation of Related Structures:
7PBC, 7PDW, 7PDX, 7QPJ - PubMed Abstract:
T-cell receptor (TCR) immunotherapy is becoming a viable modality in cancer treatment with efficacy in clinical trials. The safety of patients is paramount, so innovative cell engineering methods are being employed to exploit adaptive immunity while controlling the factors governing antigen receptor (ie, TCR) specificity and cross-reactivity. We recently reported a TCR engineering campaign and selectivity profiling assay (X-scan) targeting a melanoma antigen gene (MAGE)-A10 peptide. This helped to distinguish between two well-performing TCRs based on cross-reactivity potential during preclinical drug evaluation, allowing one to be advanced to T-cell immunotherapeutic clinical trials. Here, we present three-dimensional structural information on those TCRs, highlighting engineering improvements and molecular mechanisms likely underpinning differential selectivity. Parental and engineered TCRs were purified and crystallized either alone or complexed to human leucocyte antigen (HLA)-A*02:01 presenting the MAGE-A10 9-mer peptide, GLYDGMEHL (pHLA/MAGE-A10-9). Using X-ray diffraction, we solved four high-resolution crystal structures and evaluated them relative to previously reported functional results. The unligated parental TCR displayed similar complementarity-determining region (CDR) loop conformations when bound to pHLA/MAGE-A10-9; a rigid-body movement of TCR beta chain variable domain (TRBV) relative to TCR alpha chain variable domain helped optimal pHLA engagement. This first view of an HLA-bound MAGE-A10 peptide revealed an intrachain non-covalent 'staple' between peptide Tyr3 and Glu7. A subtle Glu31-Asp mutation in βCDR1 of the parental TCR generated a high-affinity derivative. Its pHLA-complexed structure shows that the shorter Asp leans toward the pHLA with resulting rigid-body TRBV shift, creating localized changes around the peptide's C-terminus. Structural comparison with a less selective TCR indicated that differential cross-reactivity to MAGE-A10 peptide variants is most readily explained by alterations in surface electrostatics, and the size and geometry of TCR-peptide interfacial cavities. Modest changes in engineered TCRs targeting MAGE-A10 produced significantly different properties. Conformational invariance of TCR and antigen peptide plus more space-filling CDR loop sequences may be desirable properties for clinically relevant TCR-pHLA systems to reduce the likelihood of structurally similar peptide mimics being tolerated by a TCR. Such properties may partially explain why the affinity-enhanced, in vitro-selected TCR has been generally well tolerated in patients.
- Adaptimmune, Abingdon, Oxfordshire, UK Philip.Simister@adaptimmune.com.
Organizational Affiliation: