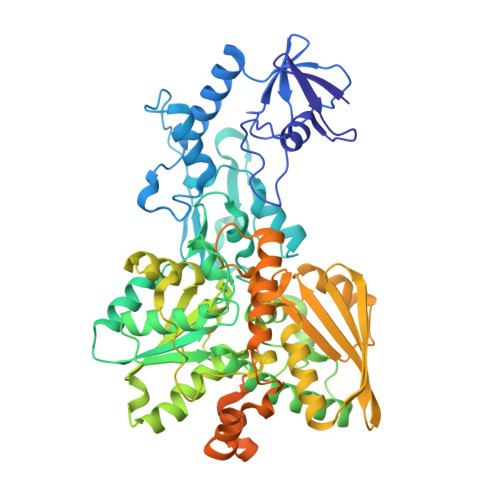
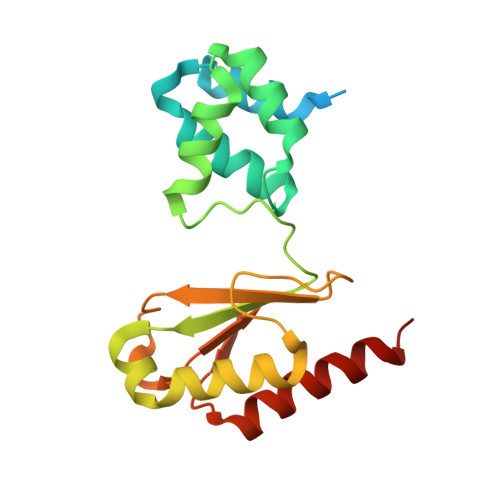
Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Furlan, C., Chongdar, N., Gupta, P., Lubitz, W., Ogata, H., Blaza, J.N., Birrell, J.A.(2022) Elife 11
- PubMed: 36018003
- DOI: https://doi.org/10.7554/eLife.79361
- Primary Citation of Related Structures:
7P5H, 7P8N, 7P91, 7P92 - PubMed Abstract:
Electron bifurcation is a fundamental energy conservation mechanism in nature in which two electrons from an intermediate-potential electron donor are split so that one is sent along a high-potential pathway to a high-potential acceptor and the other is sent along a low-potential pathway to a low-potential acceptor. This process allows endergonic reactions to be driven by exergonic ones and is an alternative, less recognized, mechanism of energy coupling to the well-known chemiosmotic principle. The electron-bifurcating [FeFe] hydrogenase from Thermotoga maritima (HydABC) requires both NADH and ferredoxin to reduce protons generating hydrogen. The mechanism of electron bifurcation in HydABC remains enigmatic in spite of intense research efforts over the last few years. Structural information may provide the basis for a better understanding of spectroscopic and functional information. Here, we present a 2.3 Å electron cryo-microscopy structure of HydABC. The structure shows a heterododecamer composed of two independent 'halves' each made of two strongly interacting HydABC heterotrimers connected via a [4Fe-4S] cluster. A central electron transfer pathway connects the active sites for NADH oxidation and for proton reduction. We identified two conformations of a flexible iron-sulfur cluster domain: a 'closed bridge' and an 'open bridge' conformation, where a Zn 2+ site may act as a 'hinge' allowing domain movement. Based on these structural revelations, we propose a possible mechanism of electron bifurcation in HydABC where the flavin mononucleotide serves a dual role as both the electron bifurcation center and as the NAD + reduction/NADH oxidation site.
- Structural Biology Laboratory and York Biomedical Research Institute, Department of Chemistry, The University of York, York, United Kingdom.
Organizational Affiliation: