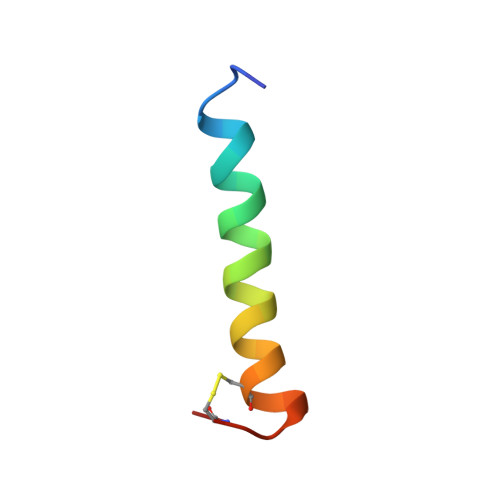
Conformation and membrane interaction studies of the potent antimicrobial and anticancer peptide palustrin-Ca.
Timmons, P.B., Hewage, C.M.(2021) Sci Rep 11: 22468-22468
- PubMed: 34789753
- DOI: https://doi.org/10.1038/s41598-021-01769-3
- Primary Citation of Related Structures:
7P4X - PubMed Abstract:
Palustrin-Ca (GFLDIIKDTGKEFAVKILNNLKCKLAGGCPP) is a host defence peptide with potent antimicrobial and anticancer activities, first isolated from the skin of the American bullfrog Lithobates catesbeianus. The peptide is 31 amino acid residues long, cationic and amphipathic. Two-dimensional NMR spectroscopy was employed to characterise its three-dimensional structure in a 50/50% water/2,2,2-trifluoroethanol-[Formula: see text] mixture. The structure is defined by an [Formula: see text]-helix that spans between Ile[Formula: see text]-Ala[Formula: see text], and a cyclic disulfide-bridged domain at the C-terminal end of the peptide sequence, between residues 23 and 29. A molecular dynamics simulation was employed to model the peptide's interactions with sodium dodecyl sulfate micelles, a widely used bacterial membrane-mimicking environment. Throughout the simulation, the peptide was found to maintain its [Formula: see text]-helical conformation between residues Ile[Formula: see text]-Ala[Formula: see text], while adopting a position parallel to the surface to micelle, which is energetically-favourable due to many hydrophobic and electrostatic contacts with the micelle.
- UCD School of Biomolecular and Biomedical Science, UCD Centre for Synthesis and Chemical Biology, UCD Conway Institute, University College Dublin, Dublin 4, Ireland. patrick.timmons@ucdconnect.ie.
Organizational Affiliation: