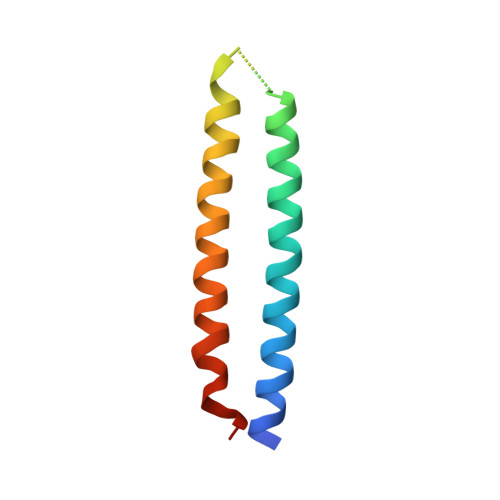
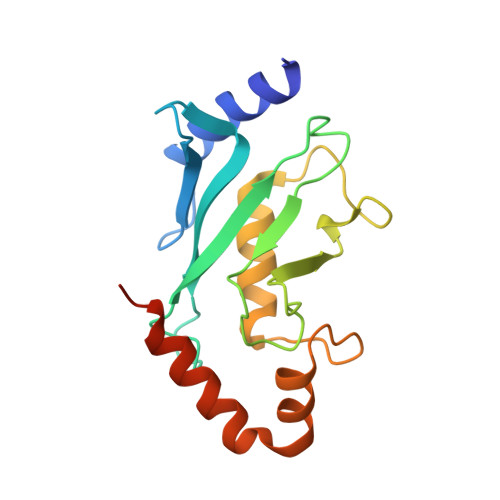
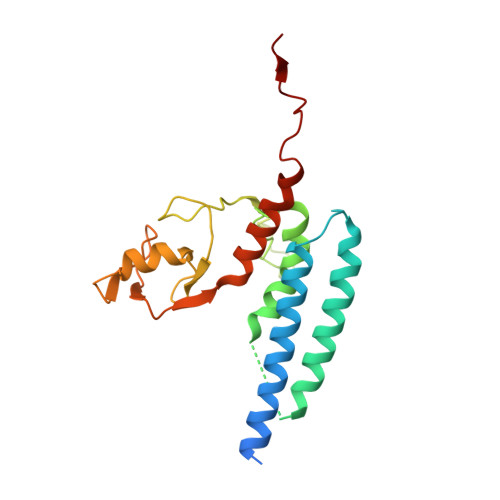
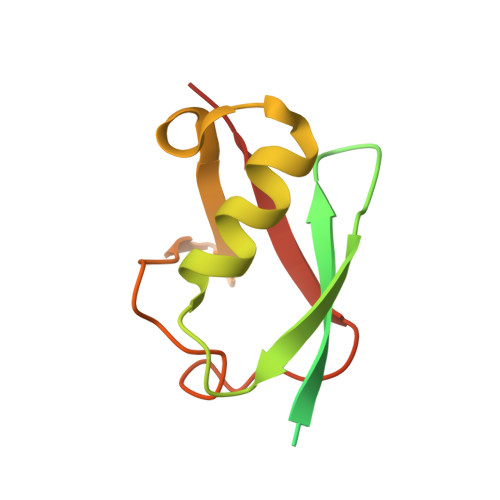
Structural basis for the E3 ligase activity enhancement of yeast Nse2 by SUMO-interacting motifs.
Varejao, N., Lascorz, J., Codina-Fabra, J., Belli, G., Borras-Gas, H., Torres-Rosell, J., Reverter, D.(2021) Nat Commun 12: 7013-7013
- PubMed: 34853311
- DOI: https://doi.org/10.1038/s41467-021-27301-9
- Primary Citation of Related Structures:
7P47 - PubMed Abstract:
Post-translational modification of proteins by ubiquitin and ubiquitin-like modifiers, such as SUMO, are key events in protein homeostasis or DNA damage response. Smc5/6 is a nuclear multi-subunit complex that participates in the recombinational DNA repair processes and is required in the maintenance of chromosome integrity. Nse2 is a subunit of the Smc5/6 complex that possesses SUMO E3 ligase activity by the presence of a SP-RING domain that activates the E2~SUMO thioester for discharge on the substrate. Here we present the crystal structure of the SUMO E3 ligase Nse2 in complex with an E2-SUMO thioester mimetic. In addition to the interface between the SP-RING domain and the E2, the complex reveals how two SIM (SUMO-Interacting Motif) -like motifs in Nse2 are restructured upon binding the donor and E2-backside SUMO during the E3-dependent discharge reaction. Both SIM interfaces are essential in the activity of Nse2 and are required to cope with DNA damage.
- Institut de Biotecnologia i de Biomedicina (IBB) and Dept. de Bioquímica i Biologia Molecular, Universitat Autònoma de Barcelona, 08193, Bellaterra, Spain.
Organizational Affiliation: