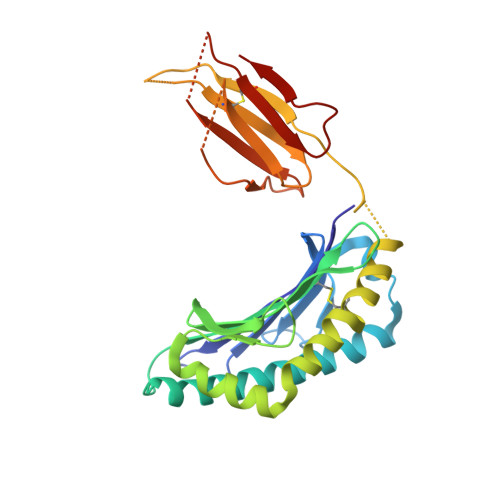
l- to d-Amino Acid Substitution in the Immunodominant LCMV-Derived Epitope gp33 Highlights the Sensitivity of the TCR Recognition Mechanism for the MHC/Peptide Structure and Dynamics.
Ballabio, F., Broggini, L., Paissoni, C., Han, X., Peqini, K., Sala, B.M., Sun, R., Sandalova, T., Barbiroli, A., Achour, A., Pellegrino, S., Ricagno, S., Camilloni, C.(2022) ACS Omega 7: 9622-9635
- PubMed: 35350306
- DOI: https://doi.org/10.1021/acsomega.1c06964
- Primary Citation of Related Structures:
7P0A, 7P0T - PubMed Abstract:
Presentation of pathogen-derived epitopes by major histocompatibility complex I (MHC-I) can lead to the activation and expansion of specific CD8 + T cell clones, eventually resulting in the destruction of infected target cells. Altered peptide ligands (APLs), designed to elicit immunogenicity toward a wild-type peptide, may affect the overall stability of MHC-I/peptide (pMHC) complexes and modulate the recognition by T cell receptors (TCR). Previous works have demonstrated that proline substitution at position 3 (p3P) of different MHC-restricted epitopes, including the immunodominant LCMV-derived epitope gp33 and escape variants, may be an effective design strategy to increase epitope immunogenicity. These studies hypothesized that the p3P substitution increases peptide rigidity, facilitating TCR binding. Here, molecular dynamics simulations indicate that the p3P modification rigidifies the APLs in solution predisposing them for the MHC-I loading as well as once bound to H-2D b , predisposing them for TCR binding. Our results also indicate that peptide position 6, key for interaction of H-2D b /gp33 with the TCR P14, takes a suboptimal conformation before as well as after binding to the TCR. Analyses of H-2D b in complex with APLs, in which position 6 was subjected to an l- to d-amino acid modification, revealed small conformational changes and comparable pMHC thermal stability. However, the l- to d-modification reduced significantly the binding to P14 even in the presence of the p3P modification. Our combined data highlight the sensitivity of the TCR for the conformational dynamics of pMHC and provide further tools to dissect and modulate TCR binding and immunogenicity via APLs.
- Dipartimento di Bioscienze, Università degli Studi di Milano, Milano 20133, Italy.
Organizational Affiliation: