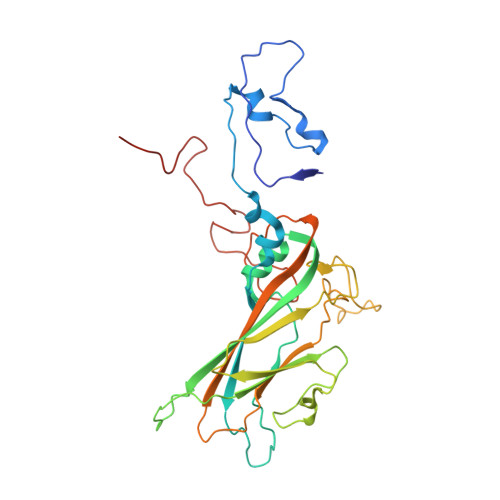
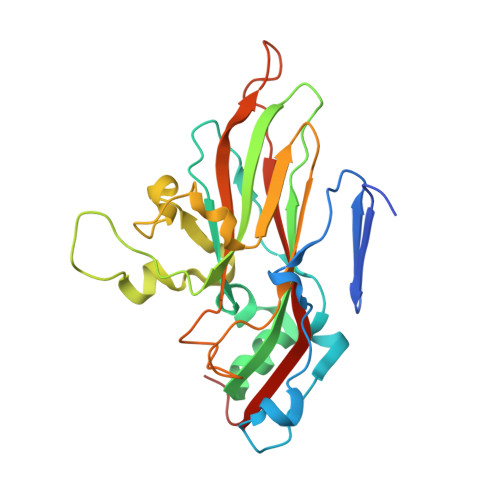
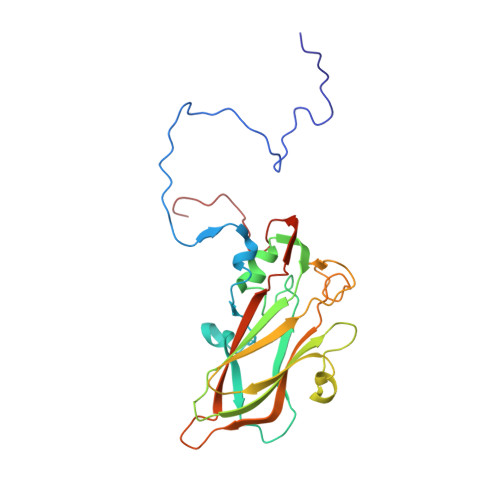
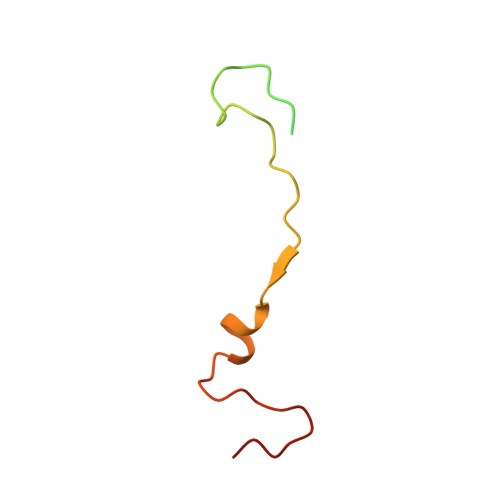
Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
Fuzik, T., Moravcova, J., Kalynych, S., Plevka, P.(2022) J Virol 96: e0060422-e0060422
- PubMed: 35939401
- DOI: https://doi.org/10.1128/jvi.00604-22
- Primary Citation of Related Structures:
7OPX, 7OZI, 7OZJ, 7OZK, 7OZL - PubMed Abstract:
Enterovirus 70 (EV70) is a human pathogen belonging to the family Picornaviridae . EV70 is transmitted by eye secretions and causes acute hemorrhagic conjunctivitis, a serious eye disease. Despite the severity of the disease caused by EV70, its structure is unknown. Here, we present the structures of the EV70 virion, altered particle, and empty capsid determined by cryo-electron microscopy. The capsid of EV70 is composed of the subunits VP1, VP2, VP3, and VP4. The partially collapsed hydrophobic pocket located in VP1 of the EV70 virion is not occupied by a pocket factor, which is commonly present in other enteroviruses. Nevertheless, we show that the pocket can be targeted by the antiviral compounds WIN51711 and pleconaril, which block virus infection. The inhibitors prevent genome release by stabilizing EV70 particles. Knowledge of the structures of complexes of EV70 with inhibitors will enable the development of capsid-binding therapeutics against this virus. IMPORTANCE Globally distributed enterovirus 70 (EV70) causes local outbreaks of acute hemorrhagic conjunctivitis. The discharge from infected eyes enables the high-efficiency transmission of EV70 in overcrowded areas with low hygienic standards. Currently, only symptomatic treatments are available. We determined the structures of EV70 in its native form, the genome release intermediate, and the empty capsid resulting from genome release. Furthermore, we elucidated the structures of EV70 in complex with two inhibitors that block virus infection, and we describe the mechanism of their binding to the virus capsid. These results enable the development of therapeutics against EV70.
- Structural Virology, Central European Institute of Technology, Masaryk University, Brno, Czech Republic.
Organizational Affiliation: