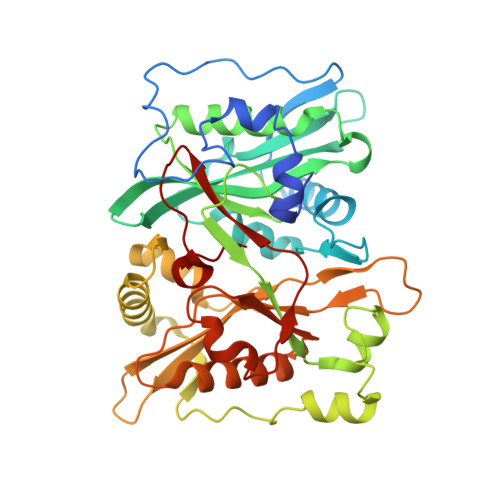
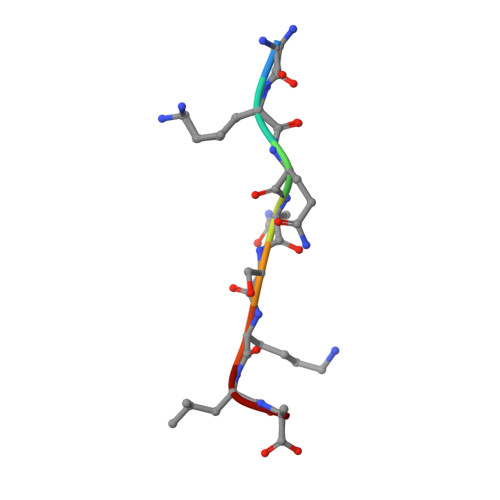
Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
Riviere, F., Dian, C., Dutheil, R.F., Monassa, P., Giglione, C., Meinnel, T.(2022) J Mol Biology 434: 167843-167843
- PubMed: 36181773
- DOI: https://doi.org/10.1016/j.jmb.2022.167843
- Primary Citation of Related Structures:
7OWM, 7OWN, 7OWO, 7OWP, 7OWQ, 7OWR, 7OWU - PubMed Abstract:
N-myristoyltransferases (NMTs) catalyze protein myristoylation, a lipid modification crucial for cell survival and a range of pathophysiological processes. Originally thought to modify only N-terminal glycine α-amino groups (G-myristoylation), NMTs were recently shown to also modify lysine ε-amino groups (K-myristoylation). However, the clues ruling NMT-dependent K-myristoylation and the full range of targets are currently unknown. Here we combine mass spectrometry, kinetic studies, in silico analysis, and crystallography to identify the specific features driving each modification. We show that direct interactions between the substrate's reactive amino group and the NMT catalytic base promote K-myristoylation but with poor efficiency compared to G-myristoylation, which instead uses a water-mediated interaction. We provide evidence of depletion of proteins with NMT-dependent K-myristoylation motifs in humans, suggesting evolutionary pressure to prevent this modification in favor of G-myristoylation. In turn, we reveal that K-myristoylation may only result from post-translational events. Our studies finally unravel the respective paths towards K-myristoylation or G-myristoylation, which rely on a very subtle tradeoff embracing the chemical landscape around the reactive group.
- Université Paris Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198 Gif-sur-Yvette cedex, France.
Organizational Affiliation: