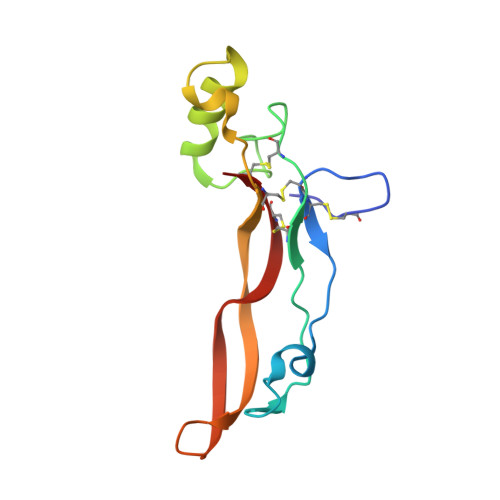
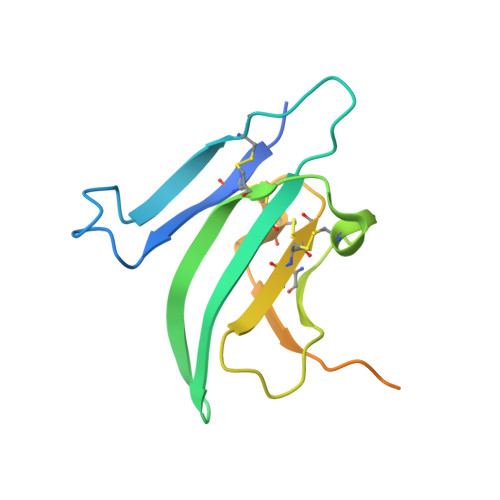
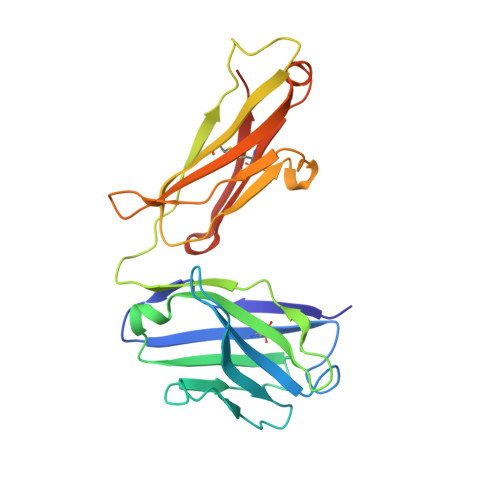
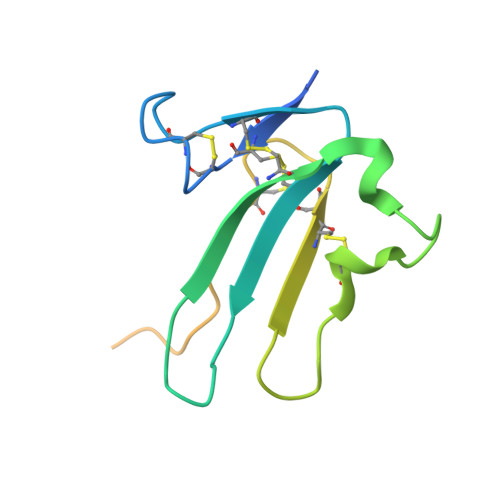
Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Goebel, E.J., Kattamuri, C., Gipson, G.R., Krishnan, L., Chavez, M., Czepnik, M., Maguire, M.C., Grenha, R., Hakansson, M., Logan, D.T., Grinberg, A.V., Sako, D., Castonguay, R., Kumar, R., Thompson, T.B.(2022) iScience 25: 103590-103590
- PubMed: 35005539
- DOI: https://doi.org/10.1016/j.isci.2021.103590
- Primary Citation of Related Structures:
7MRZ, 7OLY - PubMed Abstract:
The 30+ unique ligands of the TGFβ family signal by forming complexes using different combinations of type I and type II receptors. Therapeutically, the extracellular domain of a single receptor fused to an Fc molecule can effectively neutralize subsets of ligands. Increased ligand specificity can be accomplished by using the extracellular domains of both the type I and type II receptor to mimic the naturally occurring signaling complex. Here, we report the structure of one "type II-type I-Fc" fusion, ActRIIB-Alk4-Fc, in complex with two TGFβ family ligands, ActA, and GDF11, providing a snapshot of this therapeutic platform. The study reveals that extensive contacts are formed by both receptors, replicating the ternary signaling complex, despite the inherent low affinity of Alk4. Our study shows that low-affinity type I interactions support altered ligand specificity and can be visualized at the molecular level using this platform.
- Department of Molecular Genetics, Biochemistry, and Microbiology, University of Cincinnati, 231 Albert Sabin Way ML 0524, Cincinnati, OH 45267, USA.
Organizational Affiliation: