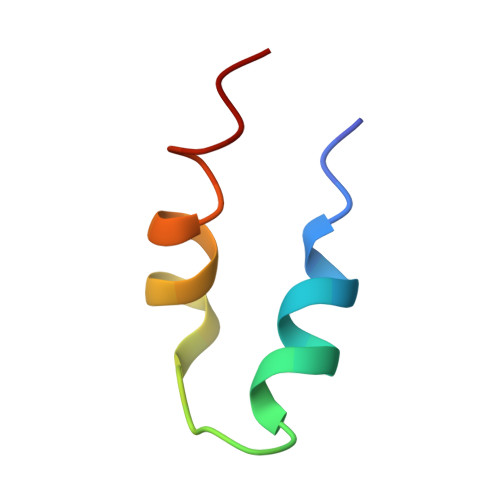
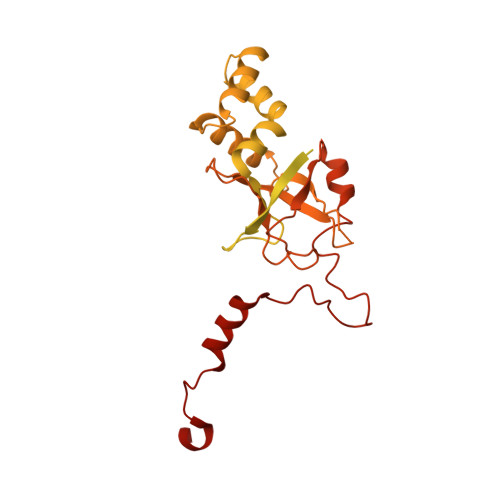
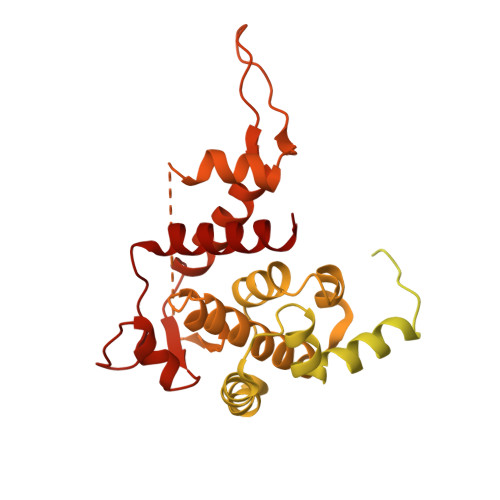
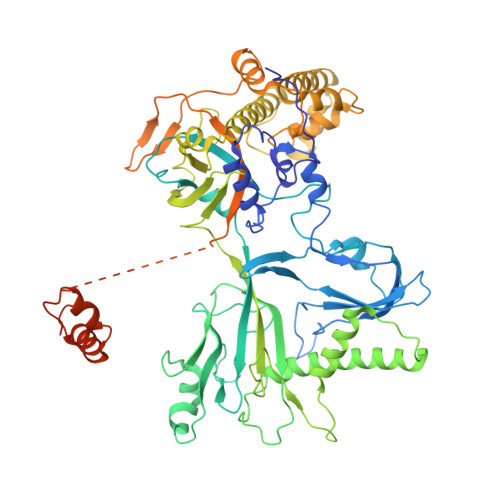
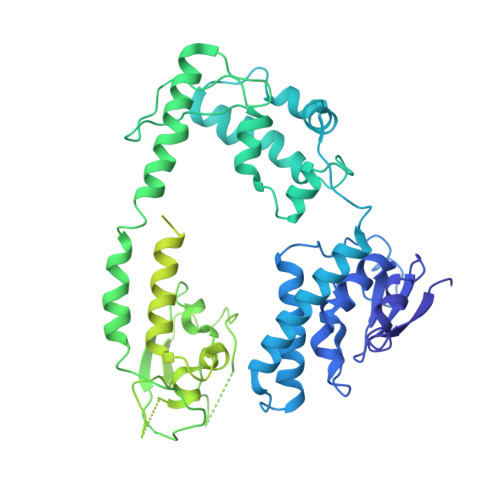
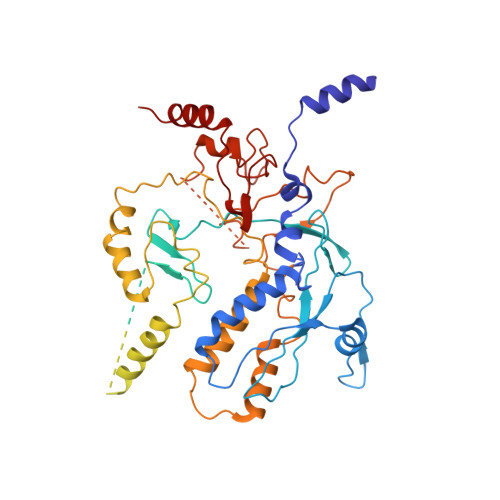
Structure of the bacteriophage PhiKZ non-virion RNA polymerase.
deYMartin Garrido, N., Orekhova, M., Lai Wan Loong, Y.T.E., Litvinova, A., Ramlaul, K., Artamonova, T., Melnikov, A.S., Serdobintsev, P., Aylett, C.H.S., Yakunina, M.(2021) Nucleic Acids Res 49: 7732-7739
- PubMed: 34181731
- DOI: https://doi.org/10.1093/nar/gkab539
- Primary Citation of Related Structures:
7OGP, 7OGR - PubMed Abstract:
Bacteriophage ΦKZ (PhiKZ) is the archetype of a family of massive bacterial viruses. It is considered to have therapeutic potential as its host, Pseudomonas aeruginosa, is an opportunistic, intrinsically antibiotic resistant, pathogen that kills tens of thousands worldwide each year. ΦKZ is an incredibly interesting virus, expressing many systems that the host already possesses. On infection, it forms a 'nucleus', erecting a barrier around its genome to exclude host endonucleases and CRISPR-Cas systems. ΦKZ infection is independent of the host transcriptional apparatus. It expresses two different multi-subunit RNA polymerases (RNAPs): the virion RNAP (vRNAP) is injected with the viral DNA during infection to transcribe early genes, including those encoding the non-virion RNAP (nvRNAP), which transcribes all further genes. ΦKZ nvRNAP is formed by four polypeptides thought to represent homologues of the eubacterial β/β' subunits, and a fifth with unclear homology, but essential for transcription. We have resolved the structure of ΦKZ nvRNAP to better than 3.0 Å, shedding light on its assembly, homology, and the biological role of the fifth subunit: it is an embedded, integral member of the complex, the position, structural homology and biochemical role of which imply that it has evolved from an ancestral homologue to σ-factor.
- Section for Structural and Synthetic Biology, Department of Infectious Disease, Imperial College London, London, UK.
Organizational Affiliation: