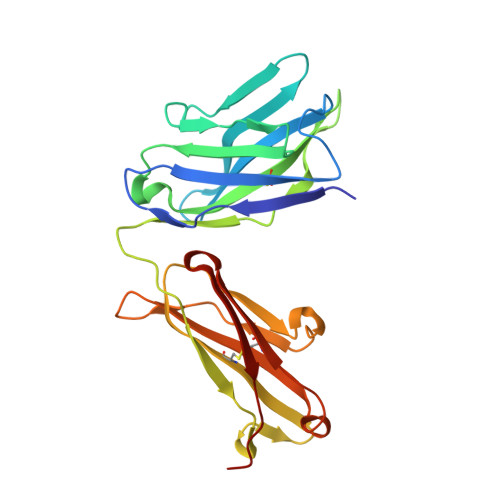
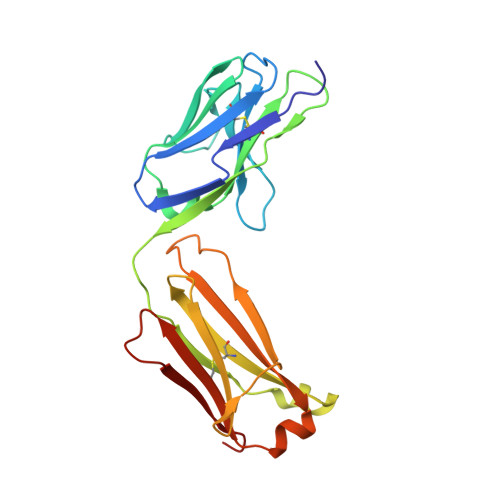
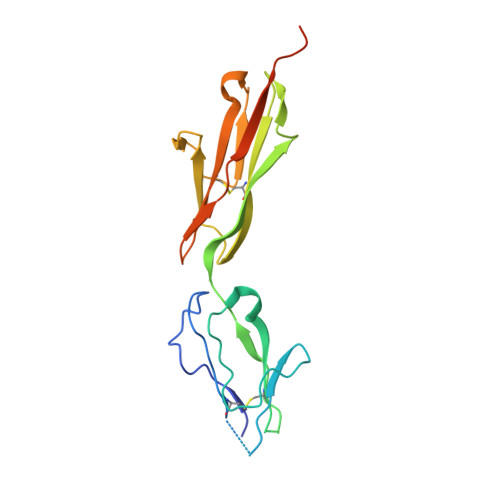
Structural details of monoclonal antibody m971 recognition of the membrane-proximal domain of CD22.
Ereno-Orbea, J., Liu, X., Sicard, T., Kucharska, I., Li, W., Borovsky, D., Cui, H., Feng, Y., Dimitrov, D.S., Julien, J.P.(2021) J Biological Chem 297: 100966-100966
- PubMed: 34273351
- DOI: https://doi.org/10.1016/j.jbc.2021.100966
- Primary Citation of Related Structures:
7O4Y, 7O52 - PubMed Abstract:
Cluster of differentiation-22 (CD22) belongs to the sialic acid-binding immunoglobulin (Ig)-like lectin family of receptors that is expressed on the surface of B cells. It has been classified as an inhibitory coreceptor for the B-cell receptor because of its function in establishing a baseline level of B-cell inhibition. The restricted expression of CD22 on B cells and its inhibitory function make it an attractive target for B-cell depletion in cases of B-cell malignancies. Genetically modified T cells with chimeric antigen receptors (CARs) derived from the m971 antibody have shown promise when used as an immunotherapeutic agent against B-cell acute lymphoblastic leukemia. A key aspect of the efficacy of this CAR-T was its ability to target a membrane-proximal epitope on the CD22 extracellular domain; however, the molecular details of m971 recognition of CD22 have thus far remained elusive. Here, we report the crystal structure of the m971 fragment antigen-binding in complex with the two most membrane-proximal Ig-like domains of CD22 (CD22 d6-d7 ). The m971 epitope on CD22 resides at the most proximal Ig domain (d7) to the membrane, and the antibody paratope contains electrostatic surfaces compatible with interactions with phospholipid head groups. Together, our data identify molecular details underlying the successful transformation of an antibody epitope on CD22 into an effective CAR immunotherapeutic target.
- Program in Molecular Medicine, The Hospital for Sick Children Research Institute, Toronto, Ontario, Canada; Ikerbasque, Basque Foundation for Science, Bilbao, Bizkaia, Spain.
Organizational Affiliation: