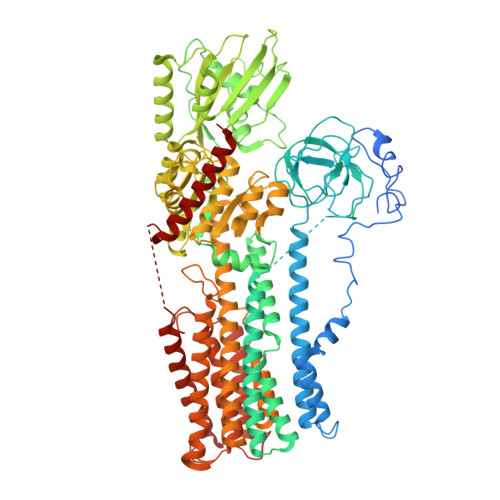
Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Heit, S., Geurts, M.M.G., Murphy, B.J., Corey, R.A., Mills, D.J., Kuhlbrandt, W., Bublitz, M.(2021) Sci Adv 7: eabj5255-eabj5255
- PubMed: 34757782
- DOI: https://doi.org/10.1126/sciadv.abj5255
- Primary Citation of Related Structures:
7NXF, 7NY1 - PubMed Abstract:
The fungal plasma membrane H + -ATPase Pma1 is a vital enzyme, generating a proton-motive force that drives the import of essential nutrients. Autoinhibited Pma1 hexamers in the plasma membrane of starving fungi are activated by glucose signaling and subsequent phosphorylation of the autoinhibitory domain. As related P-type adenosine triphosphatases (ATPases) are not known to oligomerize, the physiological relevance of Pma1 hexamers remained unknown. We have determined the structure of hexameric Pma1 from Neurospora crassa by electron cryo-microscopy at 3.3-Å resolution, elucidating the molecular basis for hexamer formation and autoinhibition and providing a basis for structure-based drug development. Coarse-grained molecular dynamics simulations in a lipid bilayer suggest lipid-mediated contacts between monomers and a substantial protein-induced membrane deformation that could act as a proton-attracting funnel.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.
Organizational Affiliation: