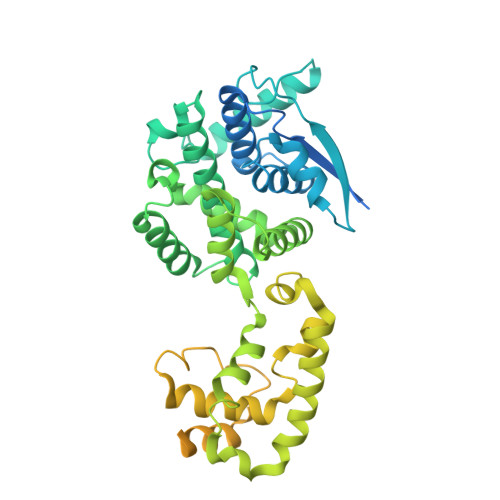
CryoEM structure of the Nipah virus nucleocapsid assembly.
Ker, D.S., Jenkins, H.T., Greive, S.J., Antson, A.A.(2021) PLoS Pathog 17: e1009740-e1009740
- PubMed: 34270629
- DOI: https://doi.org/10.1371/journal.ppat.1009740
- Primary Citation of Related Structures:
7NT5, 7NT6 - PubMed Abstract:
Nipah and its close relative Hendra are highly pathogenic zoonotic viruses, storing their ssRNA genome in a helical nucleocapsid assembly formed by the N protein, a major viral immunogen. Here, we report the first cryoEM structure for a Henipavirus RNA-bound nucleocapsid assembly, at 3.5 Å resolution. The helical assembly is stabilised by previously undefined N- and C-terminal segments, contributing to subunit-subunit interactions. RNA is wrapped around the nucleocapsid protein assembly with a periodicity of six nucleotides per protomer, in the "3-bases-in, 3-bases-out" conformation, with protein plasticity enabling non-sequence specific interactions. The structure reveals commonalities in RNA binding pockets and in the conformation of bound RNA, not only with members of the Paramyxoviridae family, but also with the evolutionarily distant Filoviridae Ebola virus. Significant structural differences with other Paramyxoviridae members are also observed, particularly in the position and length of the exposed α-helix, residues 123-139, which may serve as a valuable epitope for surveillance and diagnostics.
- York Structural Biology Laboratory, Department of Chemistry, University of York, York, United Kingdom.
Organizational Affiliation: