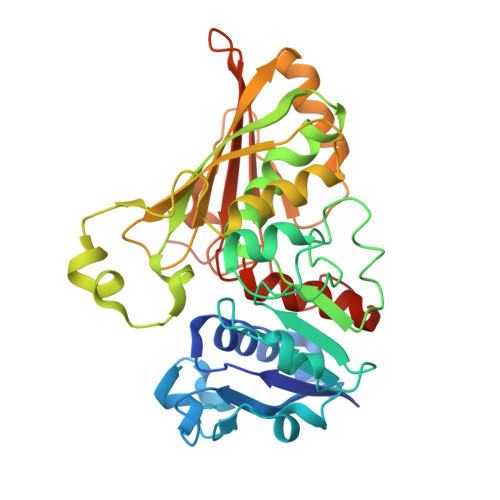
A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Gupta, P., Thomas, S.E., Zaidan, S.A., Pasillas, M.A., Cory-Wright, J., Sebastian-Perez, V., Burgess, A., Cattermole, E., Meghir, C., Abell, C., Coyne, A.G., Jacobs, W.R., Blundell, T.L., Tiwari, S., Mendes, V.(2021) Comput Struct Biotechnol J 19: 3491-3506