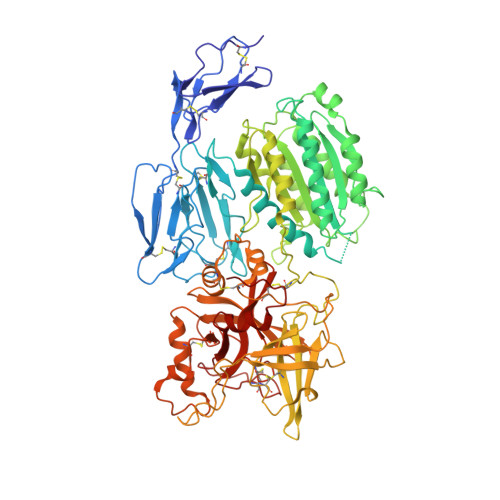
Structure determination of an unstable macromolecular complex enabled by nanobody-peptide bridging.
Lorentzen, J., Pedersen, D.V., Gadeberg, T.A.F., Andersen, G.R.(2022) Protein Sci 31: e4432-e4432
- PubMed: 36173177
- DOI: https://doi.org/10.1002/pro.4432
- Primary Citation of Related Structures:
7NOZ - PubMed Abstract:
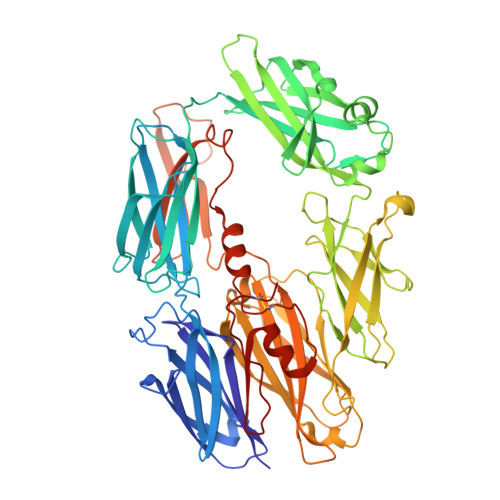
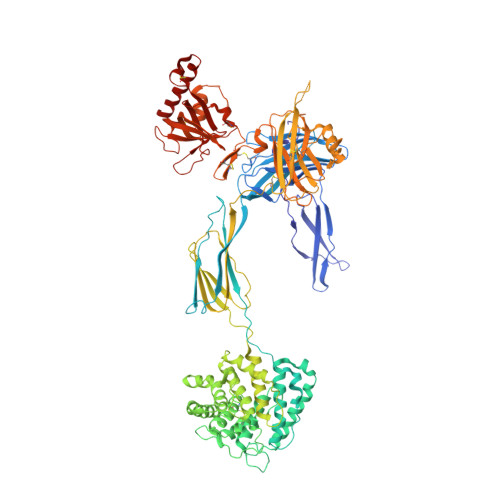
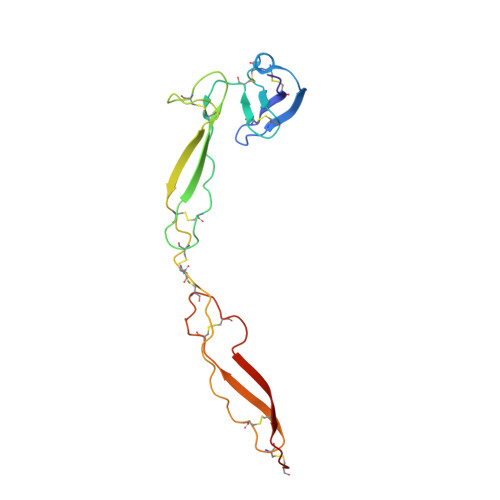
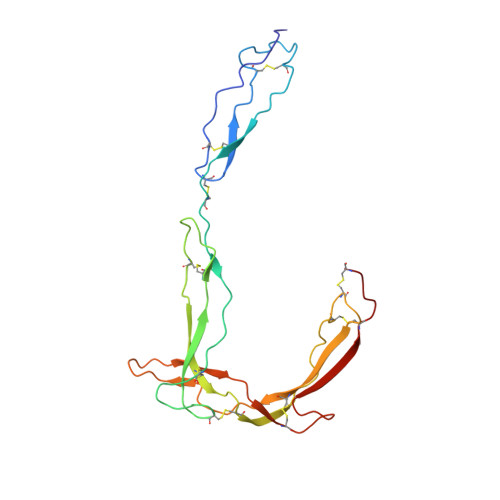
Structure determination of macromolecular complexes is challenging if subunits can dissociate during crystallization or preparation of electron microscopy grids. We present an approach where a labile complex is stabilized by linking subunits though introduction of a peptide tag in one subunit that is recognized by a nanobody tethered to a second subunit. This allowed crystal structure determination at 3.9 Å resolution of the highly non-globular 320 kDa proconvertase formed by complement components C3b, factor B, and properdin. Whereas the binding mode of properdin to C3b is preserved, an internal rearrangement occurs in the zymogen factor B von Willebrand domain type A domain compared to the proconvertase not bound to properdin. The structure emphasizes the role of two noncanonical loops in thrombospondin repeats 5 and 6 of properdin in augmenting the activity of the C3 convertase. We suggest that linking of subunits through peptide specific tethered nanobodies represents a simple alternative to approaches like affinity maturation and chemical cross-linking for the stabilization of large macromolecular complexes. Besides applications for structural biology, nanobody bridging may become a new tool for biochemical analysis of unstable macromolecular complexes and in vitro selection of highly specific binders for such complexes.
- Department of Molecular Biology and Genetics, Section for Protein Science, Aarhus Universitet, Aarhus, Denmark.
Organizational Affiliation: