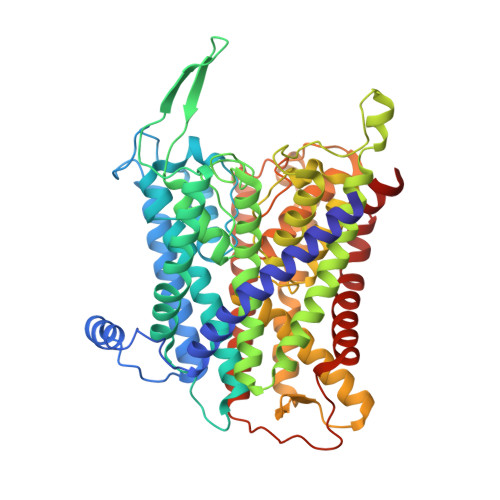
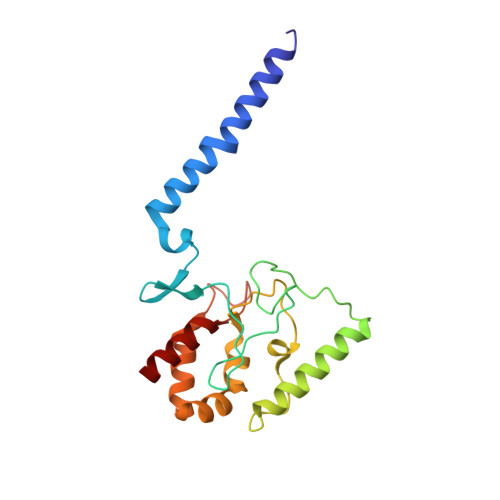
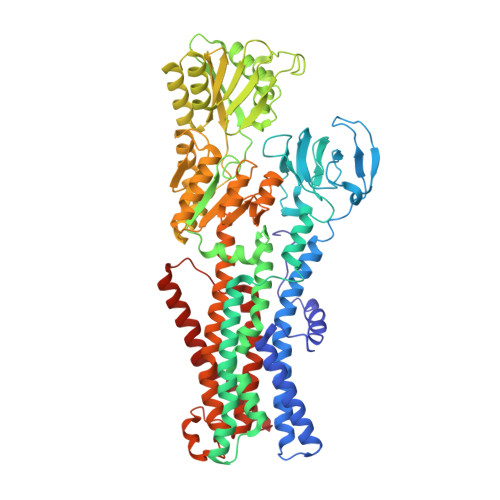
Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Silberberg, J.M., Corey, R.A., Hielkema, L., Stock, C., Stansfeld, P.J., Paulino, C., Hanelt, I.(2021) Nat Commun 12: 5098-5098
- PubMed: 34429416
- DOI: https://doi.org/10.1038/s41467-021-25242-x
- Primary Citation of Related Structures:
7NNL, 7NNP - PubMed Abstract:
KdpFABC, a high-affinity K + pump, combines the ion channel KdpA and the P-type ATPase KdpB to secure survival at K + limitation. Here, we apply a combination of cryo-EM, biochemical assays, and MD simulations to illuminate the mechanisms underlying transport and the coupling to ATP hydrolysis. We show that ions are transported via an intersubunit tunnel through KdpA and KdpB. At the subunit interface, the tunnel is constricted by a phenylalanine, which, by polarized cation-π stacking, controls K + entry into the canonical substrate binding site (CBS) of KdpB. Within the CBS, ATPase coupling is mediated by the charge distribution between an aspartate and a lysine. Interestingly, individual elements of the ion translocation mechanism of KdpFABC identified here are conserved among a wide variety of P-type ATPases from different families. This leads us to the hypothesis that KdpB might represent an early descendant of a common ancestor of cation pumps.
- Institute of Biochemistry, Biocenter, Goethe University Frankfurt, Frankfurt/Main, Germany.
Organizational Affiliation: