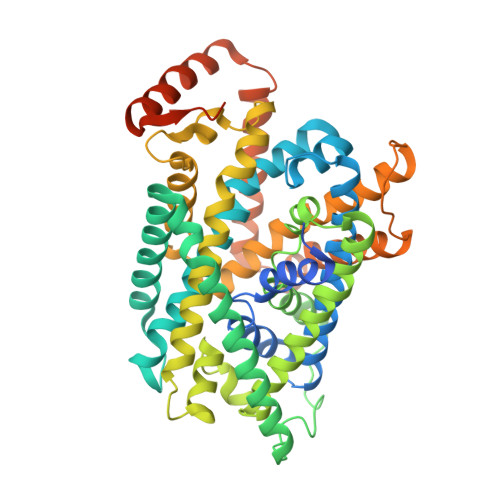
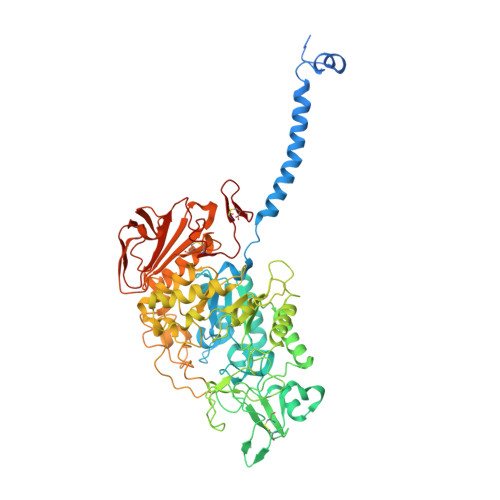
Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Lee, Y., Wiriyasermkul, P., Kongpracha, P., Moriyama, S., Mills, D.J., Kuhlbrandt, W., Nagamori, S.(2022) Nat Commun 13: 2708-2708
- PubMed: 35577790
- DOI: https://doi.org/10.1038/s41467-022-30293-9
- Primary Citation of Related Structures:
7NF6, 7NF7, 7NF8 - PubMed Abstract:
Cystinuria is a genetic disorder characterized by overexcretion of dibasic amino acids and cystine, causing recurrent kidney stones and kidney failure. Mutations of the regulatory glycoprotein rBAT and the amino acid transporter b 0,+ AT, which constitute system b 0,+ , are linked to type I and non-type I cystinuria respectively and they exhibit distinct phenotypes due to protein trafficking defects or catalytic inactivation. Here, using electron cryo-microscopy and biochemistry, we discover that Ca 2+ mediates higher-order assembly of system b 0,+ . Ca 2+ stabilizes the interface between two rBAT molecules, leading to super-dimerization of b 0,+ AT-rBAT, which in turn facilitates N-glycan maturation and protein trafficking. A cystinuria mutant T216M and mutations of the Ca 2+ site of rBAT cause the loss of higher-order assemblies, resulting in protein trapping at the ER and the loss of function. These results provide the molecular basis of system b 0,+ biogenesis and type I cystinuria and serve as a guide to develop new therapeutic strategies against it. More broadly, our findings reveal an unprecedented link between transporter oligomeric assembly and protein-trafficking diseases.
- Department of Structural Biology, Max Planck Institute of Biophysics, 60438, Frankfurt, Germany. yongchan.lee@biophys.mpg.de.
Organizational Affiliation: