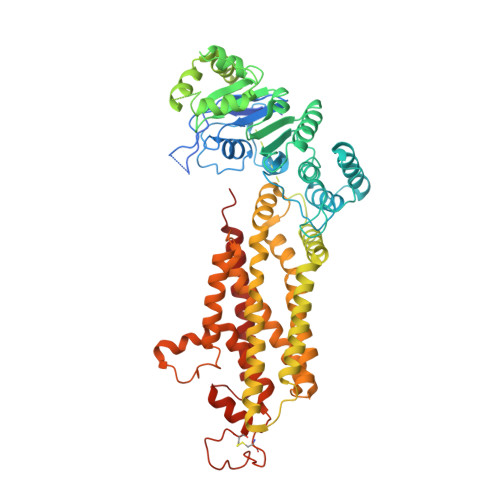
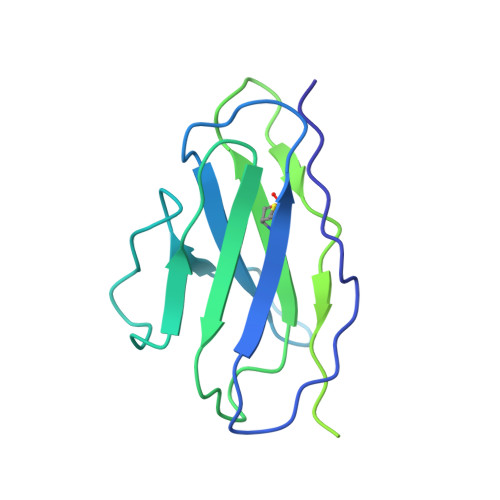
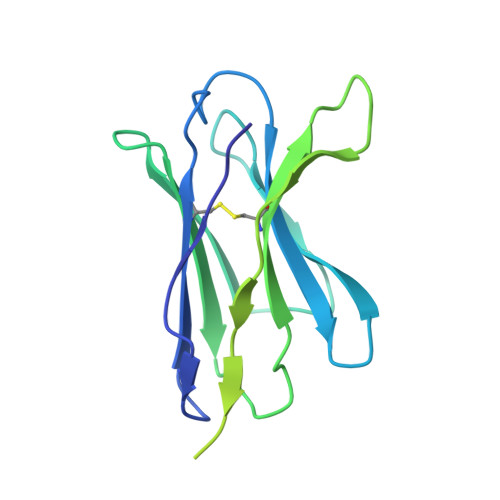
Structural Basis of Drug Recognition by the Multidrug Transporter ABCG2.
Kowal, J., Ni, D., Jackson, S.M., Manolaridis, I., Stahlberg, H., Locher, K.P.(2021) J Mol Biology 433: 166980-166980
- PubMed: 33838147
- DOI: https://doi.org/10.1016/j.jmb.2021.166980
- Primary Citation of Related Structures:
7NEQ, 7NEZ, 7NFD - PubMed Abstract:
ABCG2 is an ATP-binding cassette (ABC) transporter whose function affects the pharmacokinetics of drugs and contributes to multidrug resistance of cancer cells. While its interaction with the endogenous substrate estrone-3-sulfate (E 1 S) has been elucidated at a structural level, the recognition and recruitment of exogenous compounds is not understood at sufficiently high resolution. Here we present three cryo-EM structures of nanodisc-reconstituted, human ABCG2 bound to anticancer drugs tariquidar, topotecan and mitoxantrone. To enable structural insight at high resolution, we used Fab fragments of the ABCG2-specific monoclonal antibody 5D3, which binds to the external side of the transporter but does not interfere with drug-induced stimulation of ATPase activity. We observed that the binding pocket of ABCG2 can accommodate a single tariquidar molecule in a C-shaped conformation, similar to one of the two tariquidar molecules bound to ABCB1, where tariquidar acts as an inhibitor. We also found single copies of topotecan and mitoxantrone bound between key phenylalanine residues. Mutagenesis experiments confirmed the functional importance of two residues in the binding pocket, F439 and N436. Using 3D variability analyses, we found a correlation between substrate binding and reduced dynamics of the nucleotide binding domains (NBDs), suggesting a structural explanation for drug-induced ATPase stimulation. Our findings provide additional insight into how ABCG2 differentiates between inhibitors and substrates and may guide a rational design of new modulators and substrates.
- Institute of Molecular Biology and Biophysics, ETH Zurich, Switzerland. Electronic address: julia.kowal@mol.biol.ethz.ch.
Organizational Affiliation: