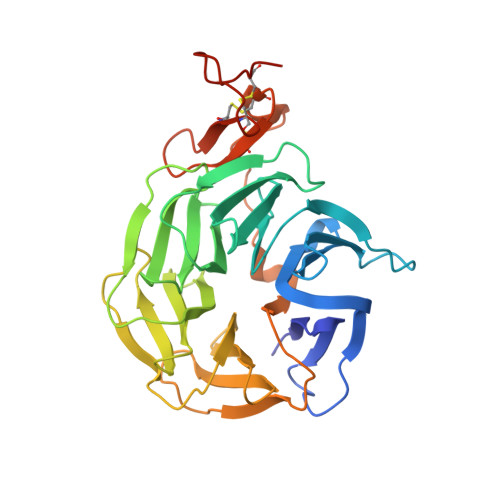
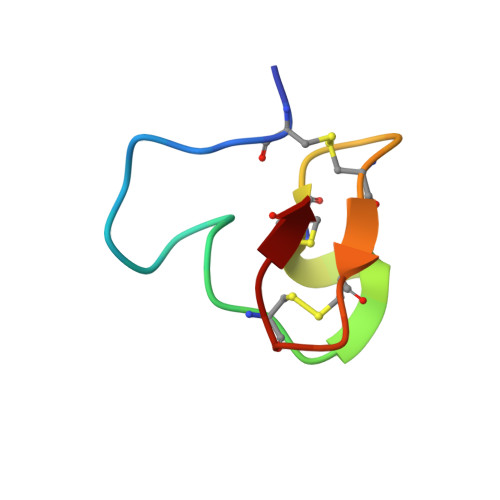
Directed evolution identifies high-affinity cystine-knot peptide agonists and antagonists of Wnt/ beta-catenin signaling.
Hansen, S., Zhang, Y., Hwang, S., Nabhan, A., Li, W., Fuhrmann, J., Kschonsak, Y., Zhou, L., Nile, A.H., Gao, X., Piskol, R., de Sousa E Melo, F., de Sauvage, F.J., Hannoush, R.N.(2022) Proc Natl Acad Sci U S A 119: e2207327119-e2207327119
- PubMed: 36343233
- DOI: https://doi.org/10.1073/pnas.2207327119
- Primary Citation of Related Structures:
7NAM - PubMed Abstract:
Developing peptide-based tools to fine-tune growth signaling pathways, in particular molecules with exquisite selectivity and high affinities, opens up opportunities for cellular reprogramming in tissue regeneration. Here, we present a library based on cystine-knot peptides (CKPs) that incorporate multiple loops for randomization and selection via directed evolution. Resulting binders could be assembled into multimeric structures to fine-tune cellular signaling. An example is presented for the Wnt pathway, which plays a key role in the homeostasis and regeneration of tissues such as lung, skin, and intestine. We discovered picomolar affinity CKP agonists of the human LPR6 receptor by exploring the limits of the topological manipulation of LRP6 dimerization. Structural analyses revealed that the agonists bind at the first β-propeller domain of LRP6, mimicking the natural Wnt inhibitors DKK1 and SOST. However, the CKP agonists exhibit a different mode of action as they amplify the signaling of natural Wnt ligands but do not activate the pathway by themselves. In an alveolosphere organoid model, the CKP agonists induced alveolar stem cell activity. They also stimulated growth in primary human intestinal organoids. The approach described here advances the important frontier of next-generation agonist design and could be applied to other signaling pathways to discover tunable agonist ligands.
- Department of Early Discovery Biochemistry, Genentech, South San Francisco, CA.
Organizational Affiliation: