CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
Gerber, E.E., Digianantonio, K.M., Tripler, T.N., Smaga, S.S., Summers, B.J., Xiong, Y.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Capsid protein | A [auth BBB], B [auth CCC], C [auth AAA] | 222 | Human immunodeficiency virus 1 | Mutation(s): 0 Gene Names: gag | 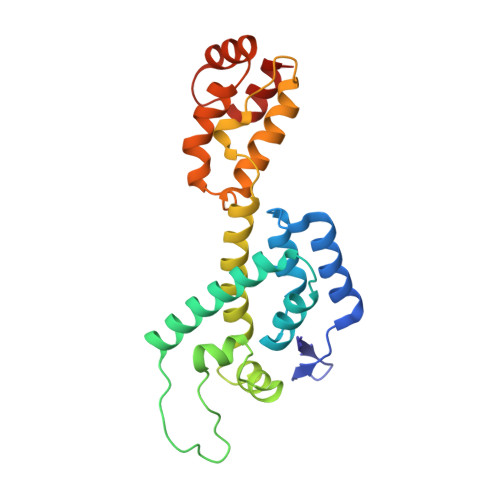 |
UniProt | |||||
Find proteins for B6DRA0 (Human immunodeficiency virus type 1) Explore B6DRA0 Go to UniProtKB: B6DRA0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B6DRA0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody | D [auth FFF] | 115 | Lama glama | Mutation(s): 0 | 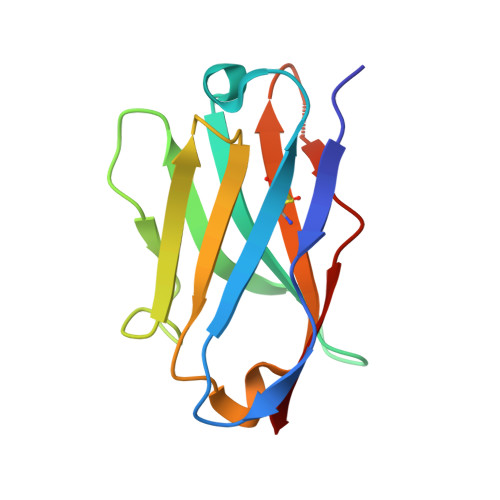 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody | E [auth EEE] | 112 | Lama glama | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody | F [auth DDD] | 114 | Lama glama | Mutation(s): 0 | 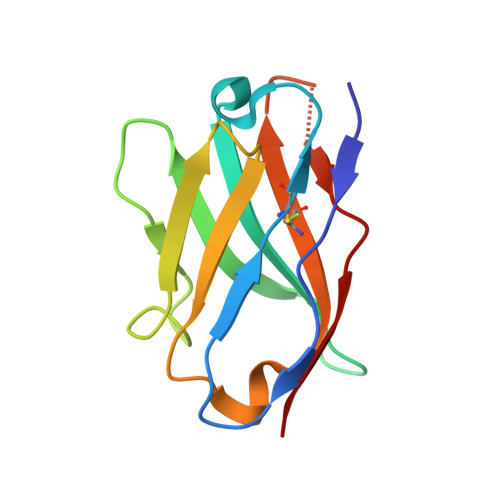 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptidyl-prolyl cis-trans isomerase A | G [auth GGG], H [auth HHH], I [auth III] | 165 | Homo sapiens | Mutation(s): 0 Gene Names: PPIA, CYPA EC: 5.2.1.8 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62937 (Homo sapiens) Explore P62937 Go to UniProtKB: P62937 | |||||
PHAROS: P62937 GTEx: ENSG00000196262 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62937 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 127.95 | α = 90 |
| b = 137.85 | β = 90 |
| c = 313.832 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DENZO | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | P50GM082251 |
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | F32 GM131426 01A1 |