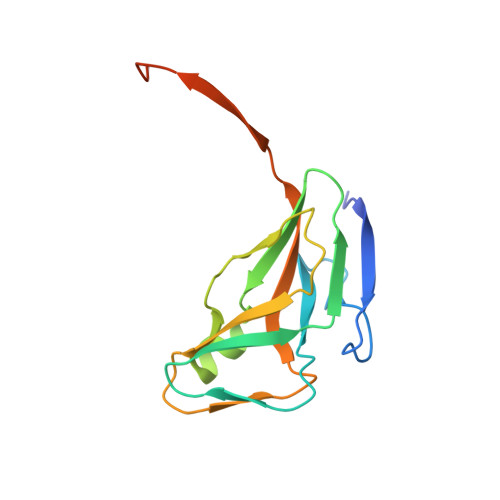
Crystal Structure of deoxyuridine 5'-triphosphate nucleotidohydrolase from Rickettsia prowazekii str. Madrid E in complex with 2'-deoxyuridine 5'-monophoephate (dUMP)
Abendroth, J., deBouver, N.D., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.